> Dataset: GSE102203
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of ARHGAP15 expression and immune infiltrating cell level correlation result in EBV+ samples
|
ARHGAP15 expression: Low -> High
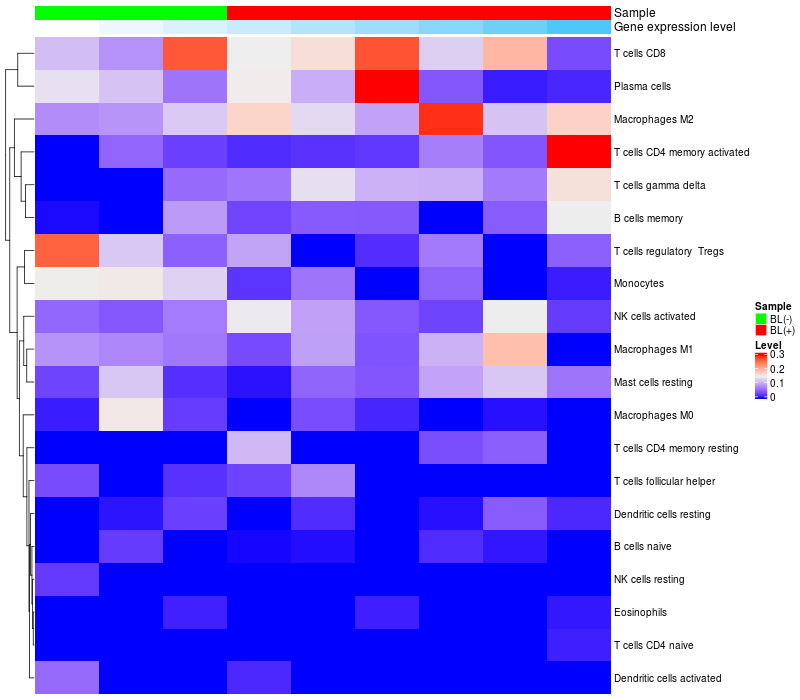
|
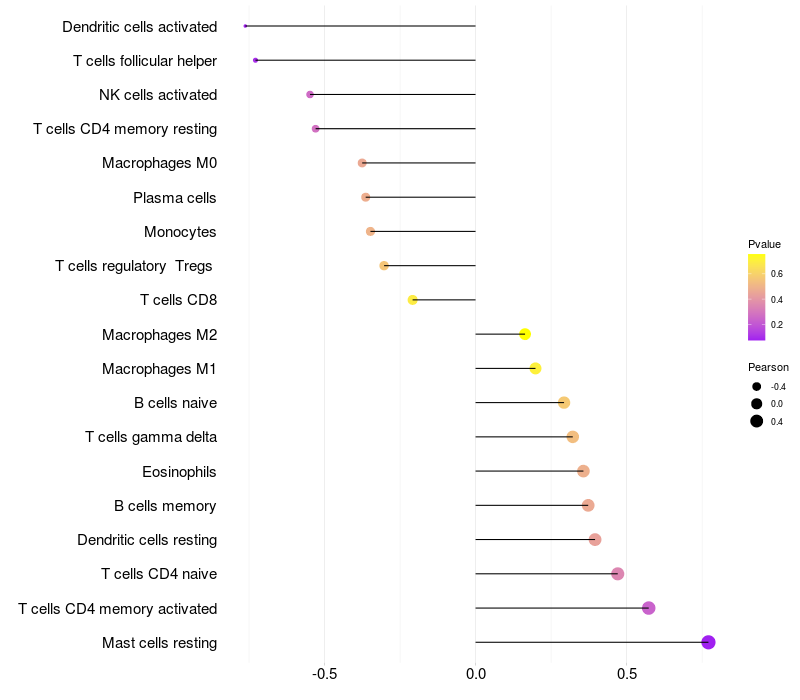
|
|
> Dataset: GSE100458
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of ARHGAP15 expression and immune infiltrating cell level correlation result in EBV+ samples
|
ARHGAP15 expression: Low -> High
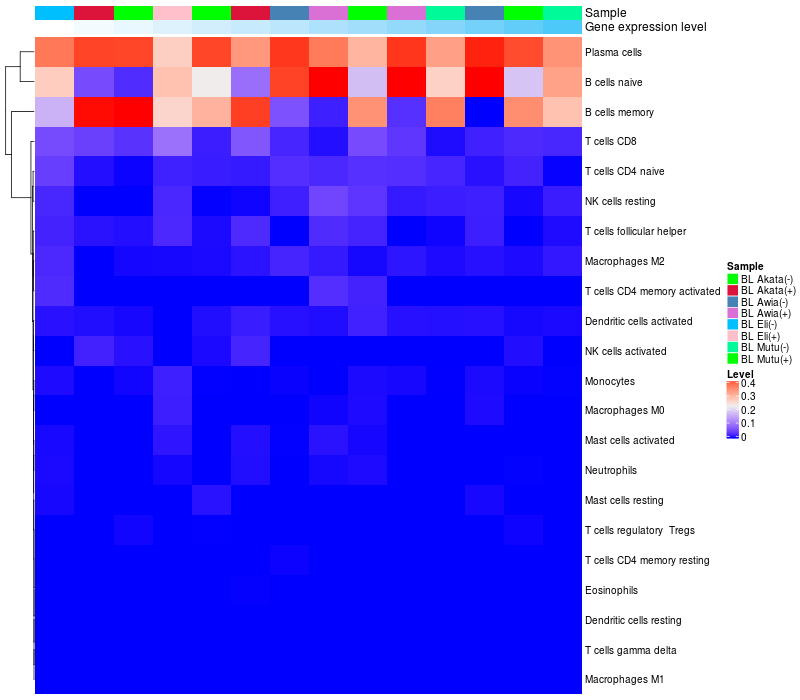
|
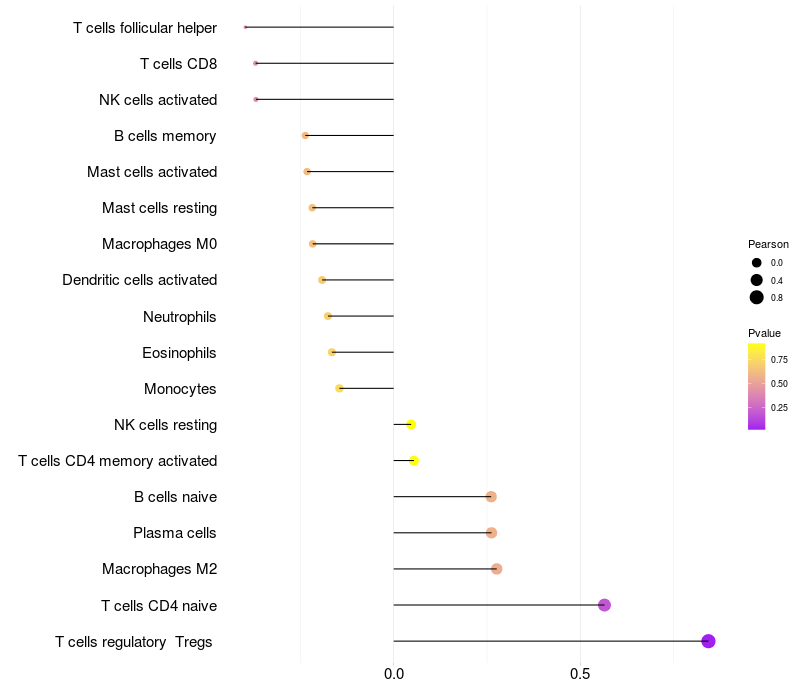
|
|
> Dataset: GSE135644
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of ARHGAP15 expression and immune infiltrating cell level correlation result in EBV+ samples
|
ARHGAP15 expression: Low -> High
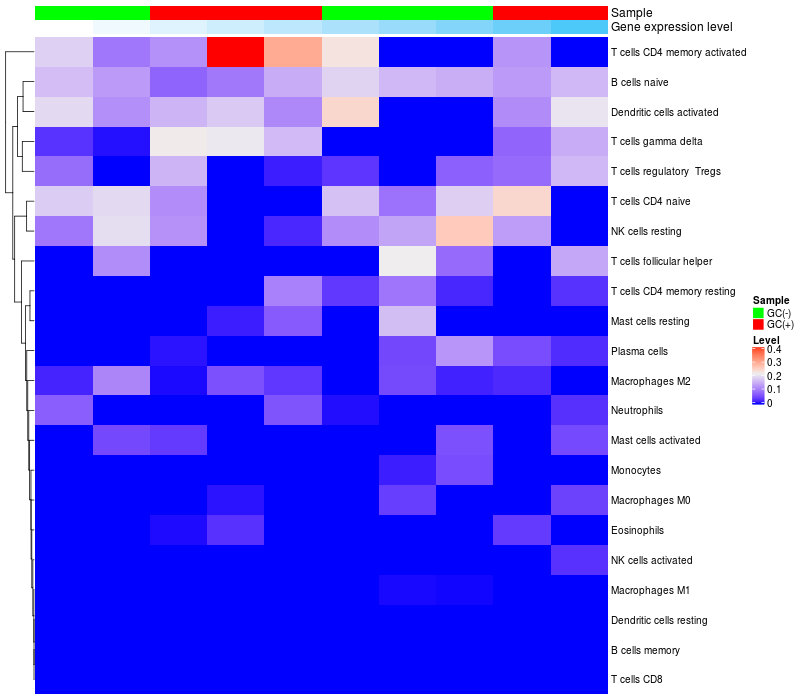
|
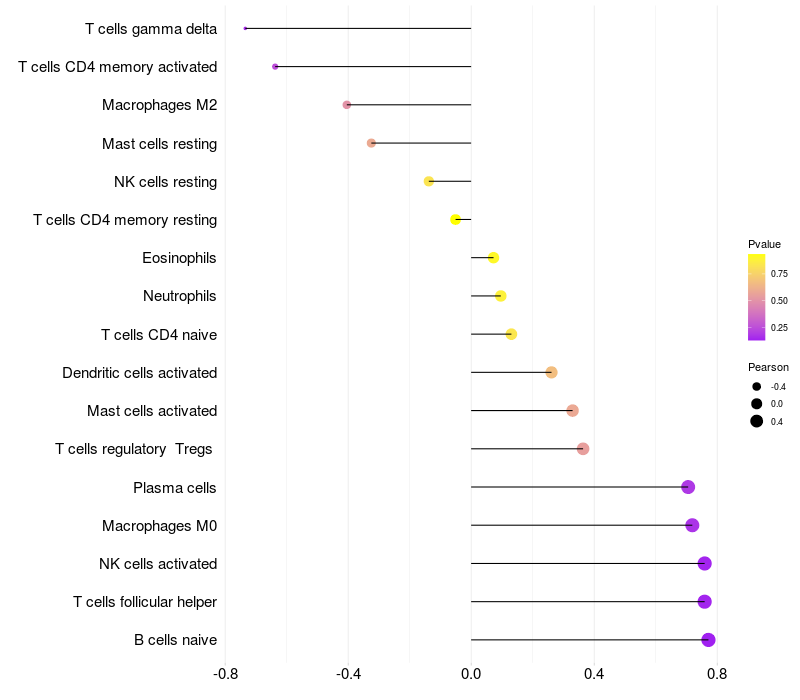
|
|
> Dataset: GSE45919
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of ARHGAP15 expression and immune infiltrating cell level correlation result in EBV+ samples
|
ARHGAP15 expression: Low -> High
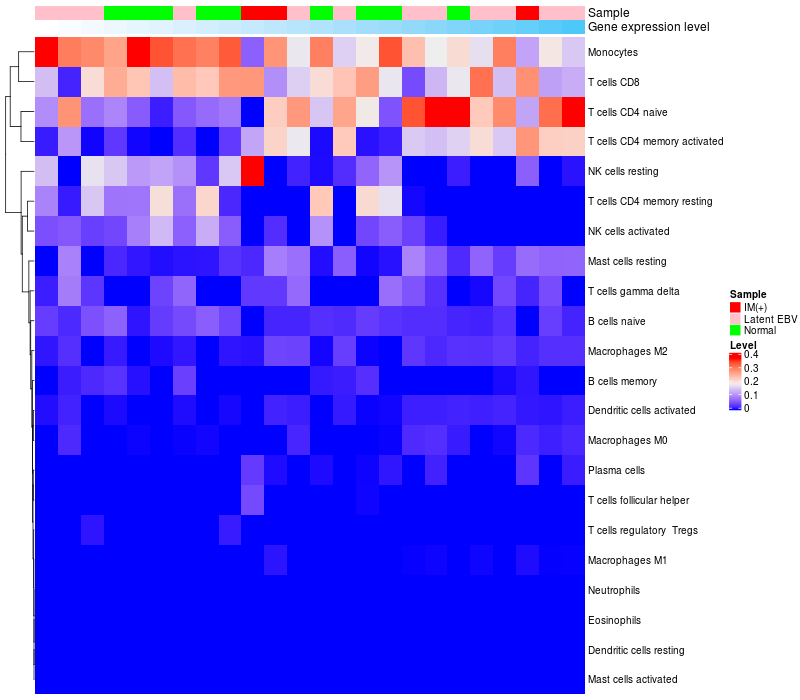
|
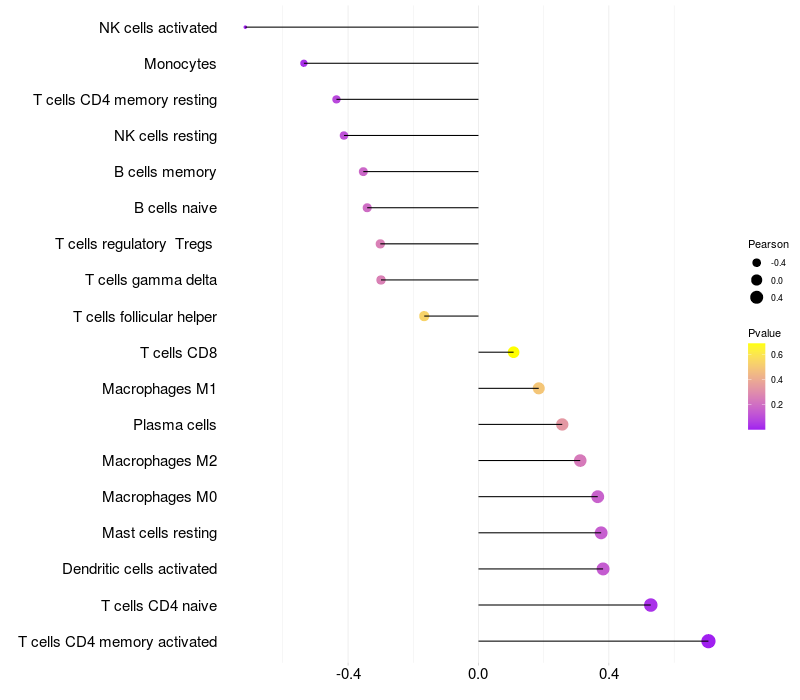
|
|
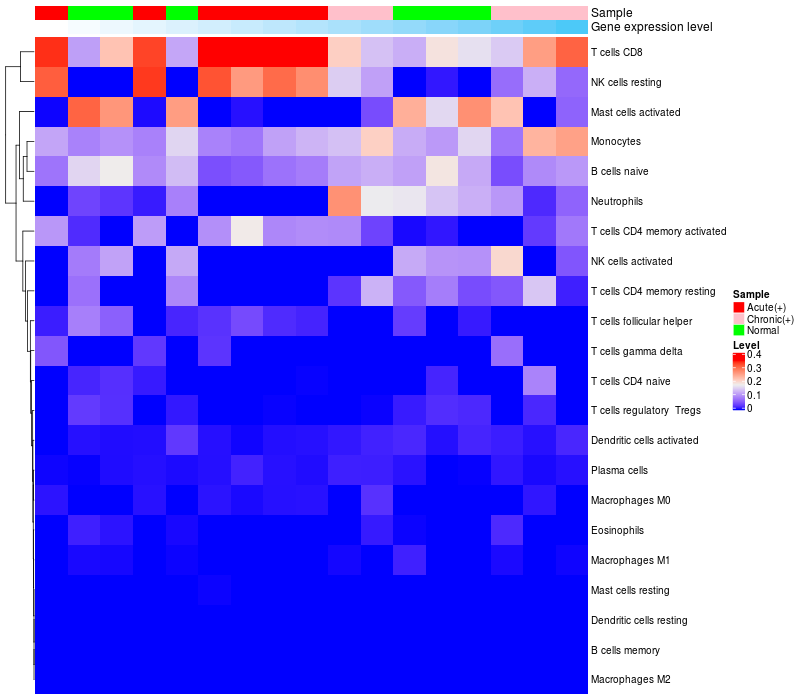
> Dataset: GSE85599
|
|
HeatMap of cell type fractions in all samples
|
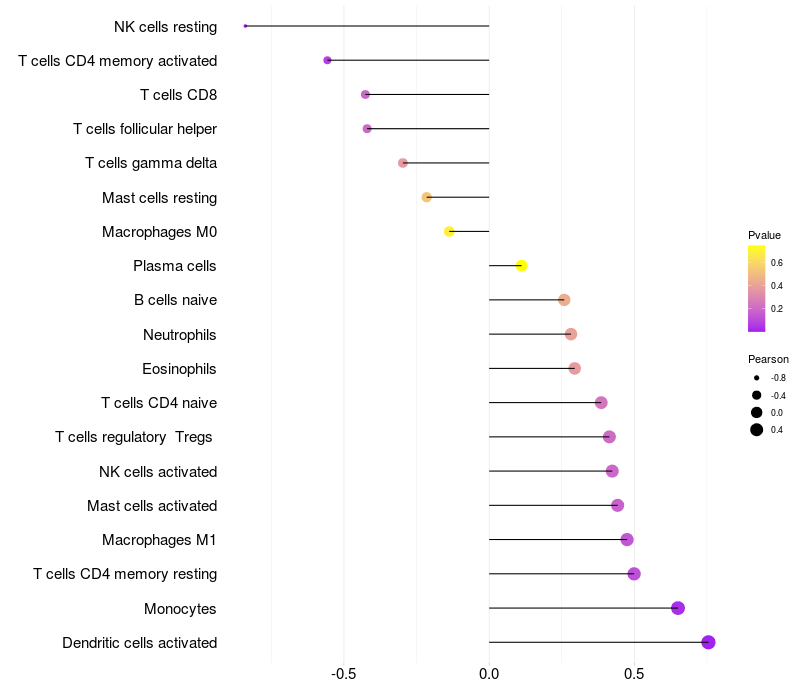
Lollipop of ARHGAP15 expression and immune infiltrating cell level correlation result in EBV+ samples
|
ARHGAP15 expression: Low -> High
|
|
|
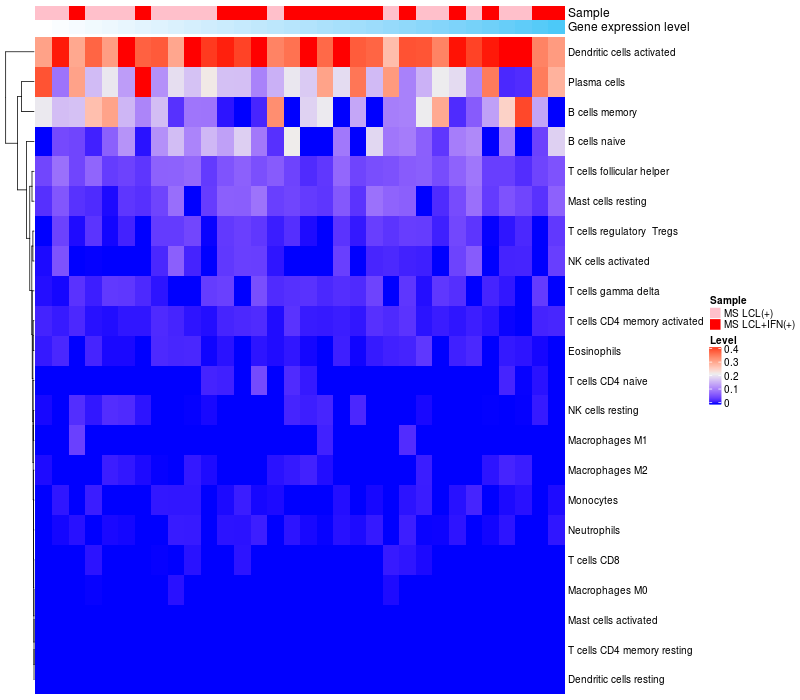
> Dataset: GSE58240
|
|
HeatMap of cell type fractions in all samples
|
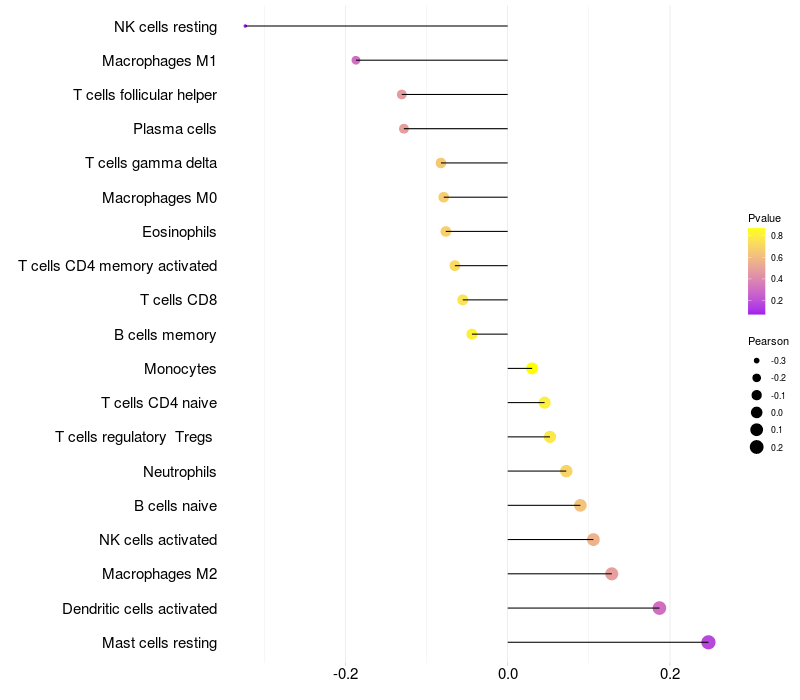
Lollipop of ARHGAP15 expression and immune infiltrating cell level correlation result in EBV+ samples
|
ARHGAP15 expression: Low -> High
|
|
|
 EBV Target gene Detail Information
EBV Target gene Detail Information