Gene Information
|
Gene Name
|
BICRA |
|
Gene ID
|
29998
|
|
Gene Full Name
|
BRD4 interacting chromatin remodeling complex associated protein |
|
Gene Alias
|
CSS12|GLTSCR1|SMARCK1 |
|
Transcripts
|
ENSG00000063169
|
|
Virus
|
HTLV1 |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Predicted intracellular proteins |
|
Uniport_ID
|
Q9NZM4
|
|
HGNC ID
|
HGNC:4332
|
|
OMIM ID
|
605690 |
|
Summary
|
Enables transcription regulator activator activity. Involved in positive regulation of DNA-templated transcription. Located in nucleus. Part of SWI/SNF complex. Implicated in Coffin-Siris syndrome 12. [provided by Alliance of Genome Resources, Apr 2025] |
Target gene [BICRA] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
Target gene [BICRA] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE52244
|
Expression array |
15 |
[HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [probe set (exon) version] |
|
GSE224047
|
RNA-seq |
10 |
Illumina NextSeq 500 (Homo sapiens) |
|
C GSE94732
|
Chip-seq |
24 |
Illumina NextSeq 500 (Homo sapiens);illumina Genome Analyzer IIx (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE94732 - BICRA peak across samples
|
Peak Plot
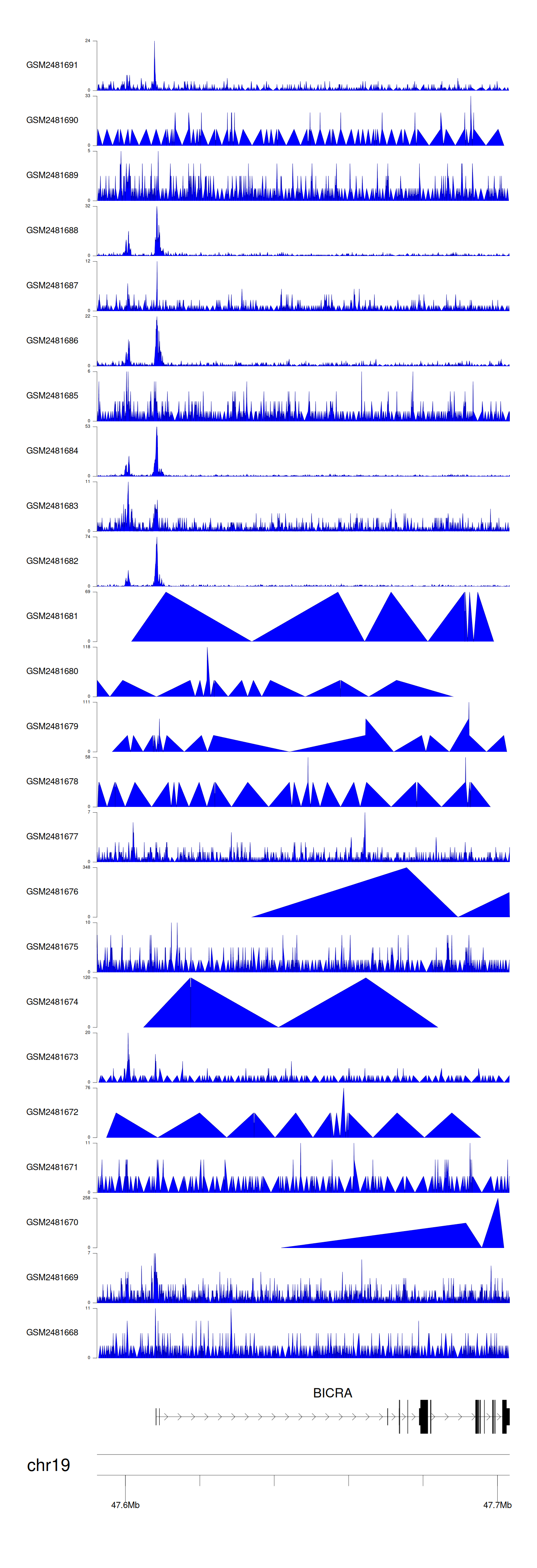
|
|
|
 HTLV1 Target gene Detail Information
HTLV1 Target gene Detail Information