Gene Information
|
Gene Name
|
DCC |
|
Gene ID
|
1630
|
|
Gene Full Name
|
DCC netrin 1 receptor |
|
Gene Alias
|
CRC18|CRCR1|HGPPS2|IGDCC1|MRMV1|NTN1R1 |
|
Transcripts
|
ENSG00000187323
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Golgi apparatus;Centriolar satellite, Centrosome, Basal body; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P43146
|
|
HGNC ID
|
HGNC:2701
|
|
OMIM ID
|
120470 |
|
Summary
|
This gene encodes a netrin 1 receptor. The transmembrane protein is a member of the immunoglobulin superfamily of cell adhesion molecules, and mediates axon guidance of neuronal growth cones towards sources of netrin 1 ligand. The cytoplasmic tail interacts with the tyrosine kinases Src and focal adhesion kinase (FAK, also known as PTK2) to mediate axon attraction. The protein partially localizes to lipid rafts, and induces apoptosis in the absence of ligand. The protein functions as a tumor suppressor, and is frequently mutated or downregulated in colorectal cancer and esophageal carcinoma. [provided by RefSeq, Oct 2009] |
Target gene [DCC] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1009657 |
chr18 |
0 |
7 |
8 |
15 |
View |
| 1012228 |
chr18 |
7 |
0 |
0 |
7 |
View |
| 1013949 |
chr18 |
2 |
0 |
2 |
4 |
View |
| 1013986 |
chr18 |
2 |
0 |
2 |
4 |
View |
| 1017151 |
chr18 |
1 |
0 |
0 |
1 |
View |
Target gene [DCC] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - DCC peak across samples
|
Peak Plot
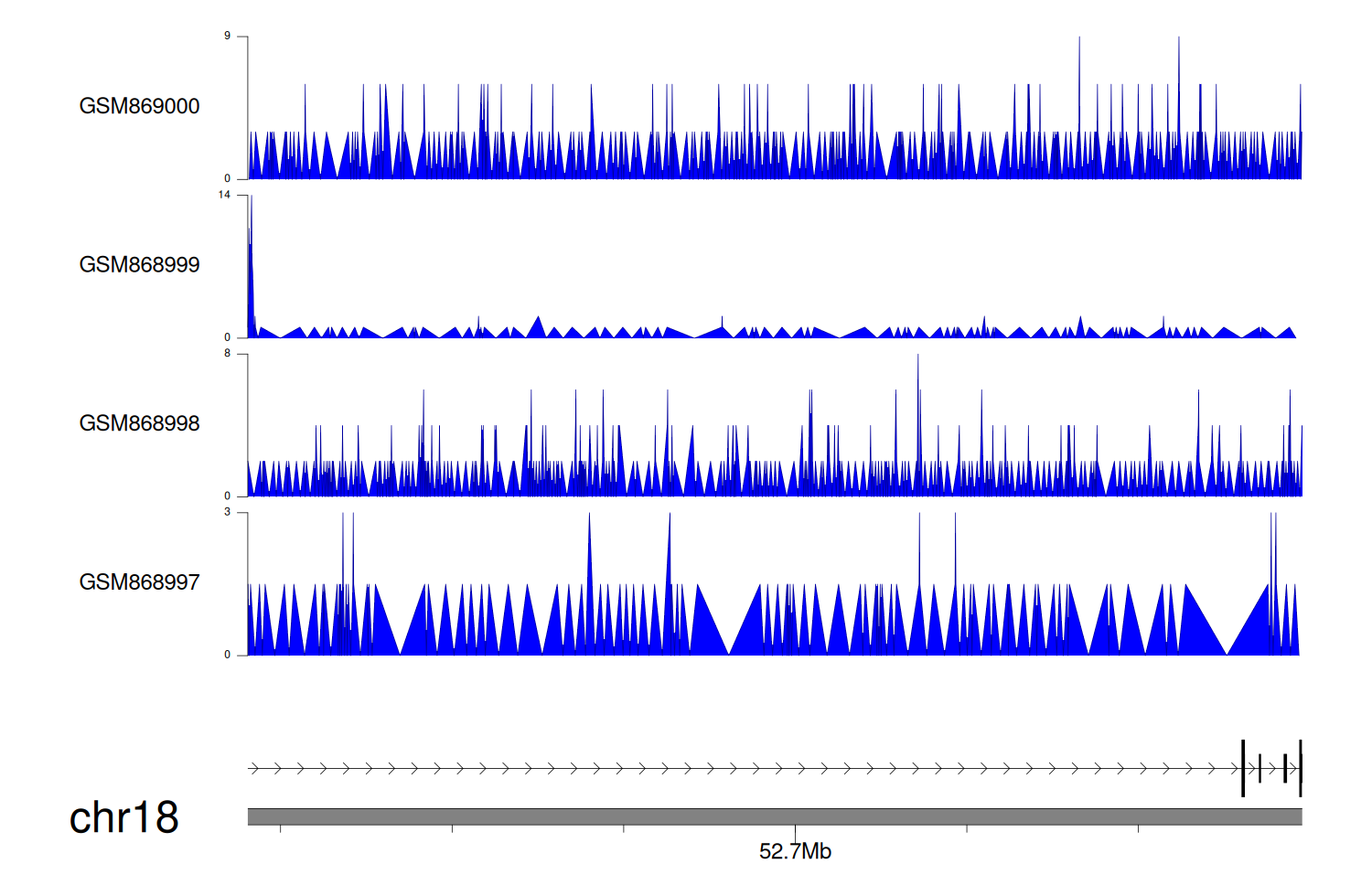
|
> Dataset: GSE68402 - DCC peak across samples
|
Peak Plot
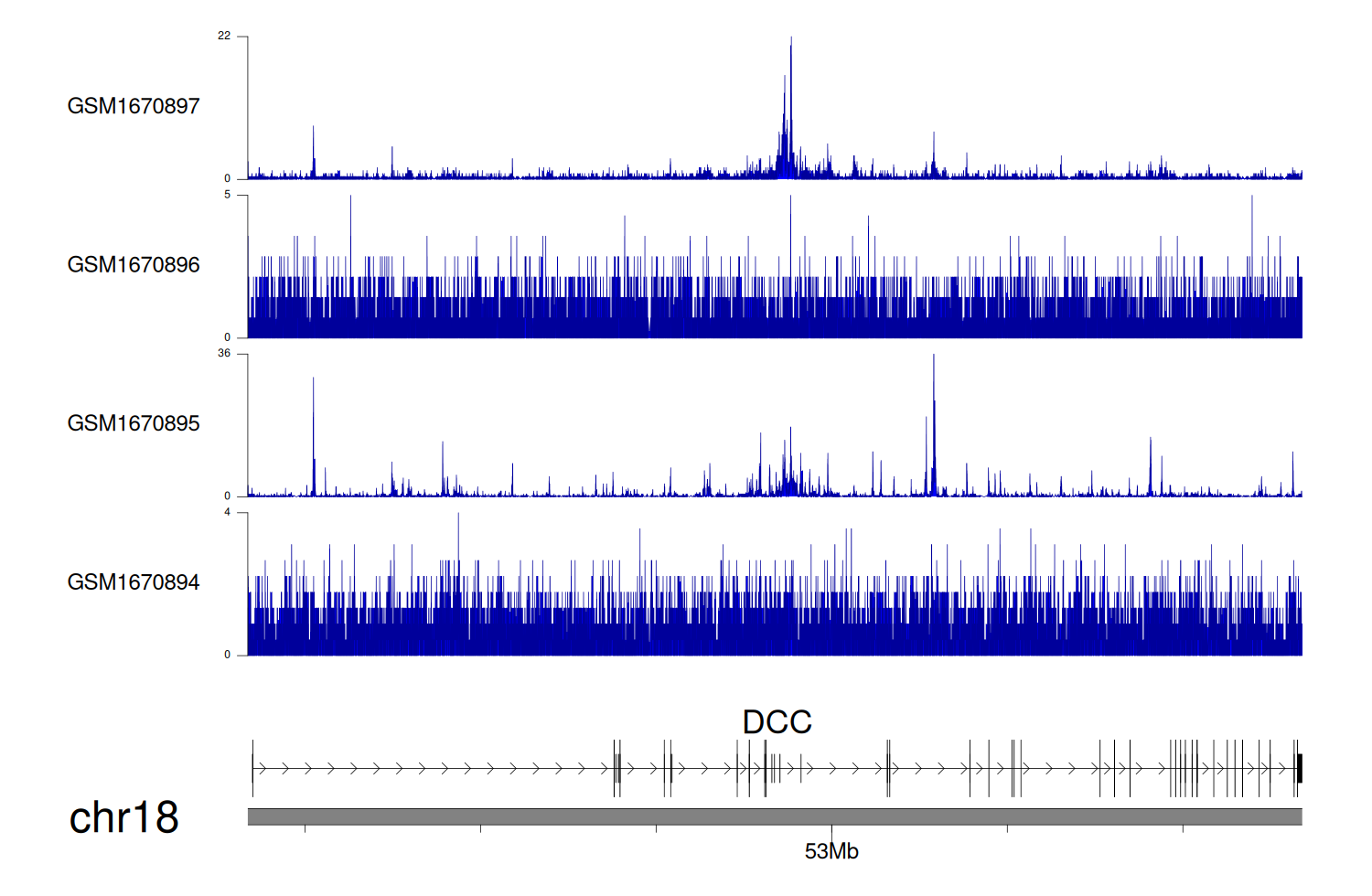
|
> Dataset: GSE270130 - DCC peak across samples
|
Peak Plot
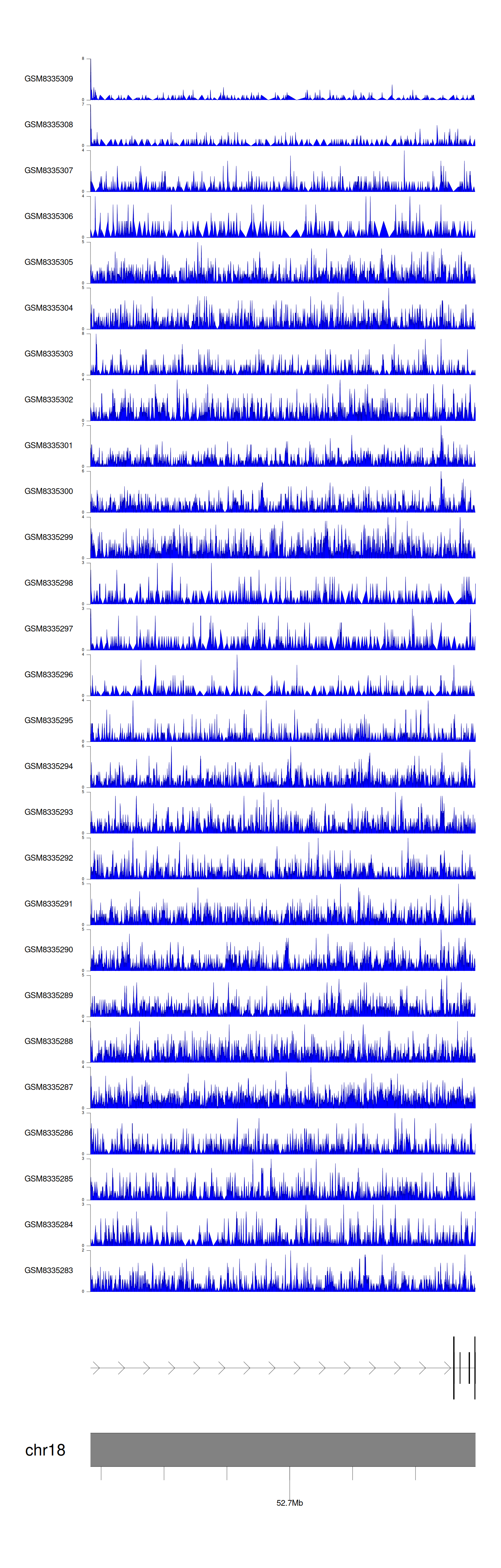
|
> Dataset: GSE100400 - DCC peak across samples
|
Peak Plot
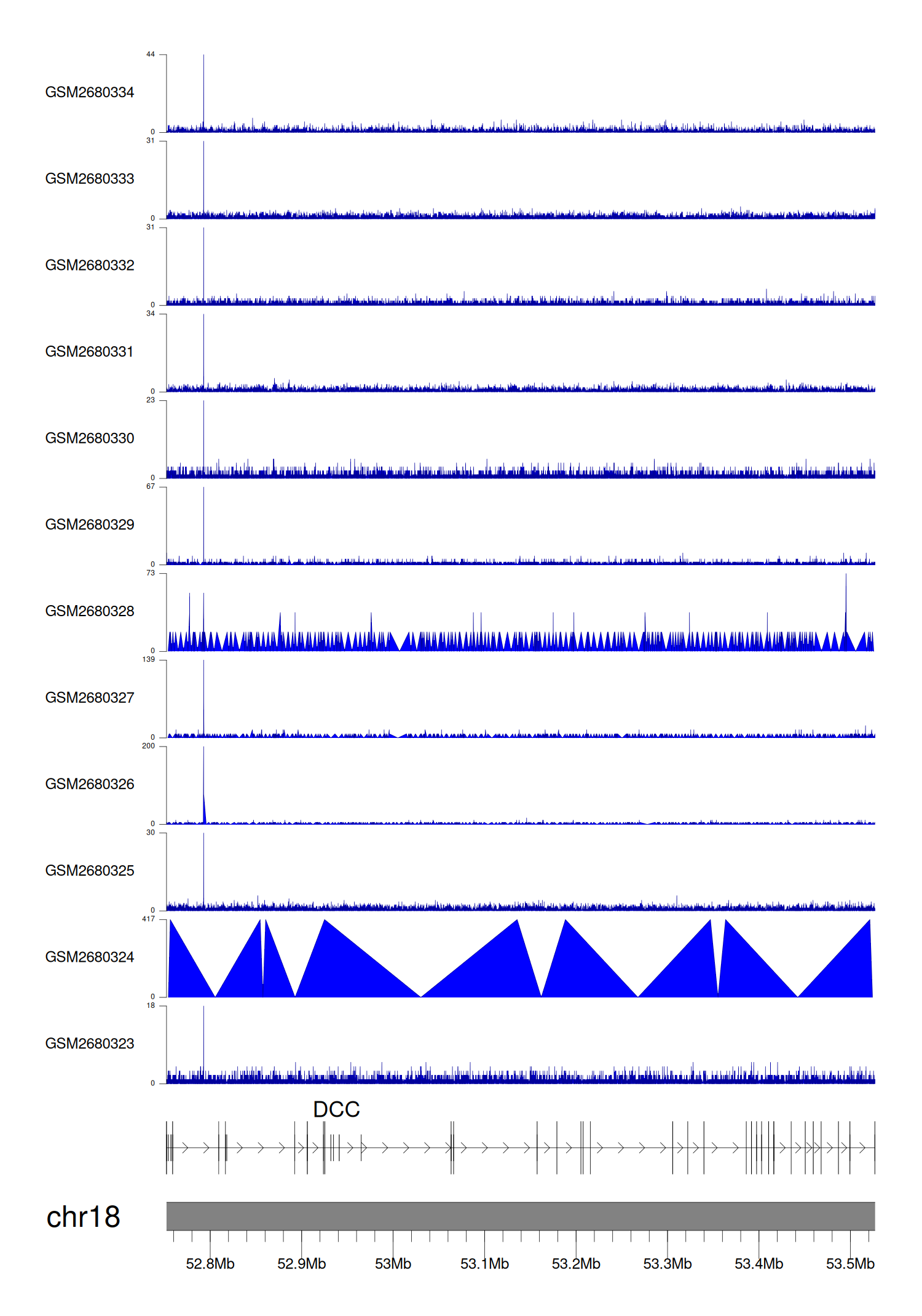
|
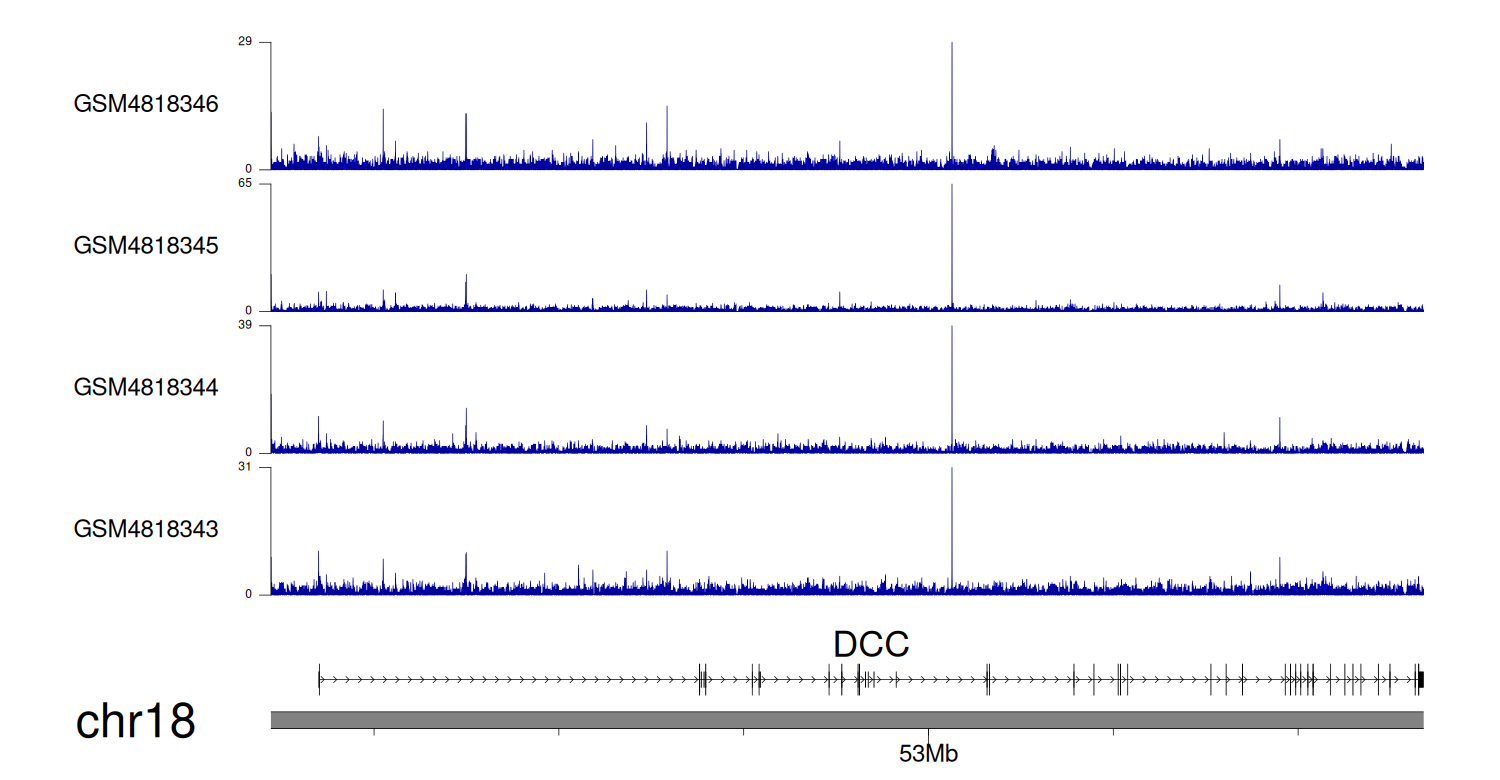
> Dataset: GSE131257 - DCC peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information