Gene Information
|
Gene Name
|
DMD |
|
Gene ID
|
1756
|
|
Gene Full Name
|
dystrophin |
|
Gene Alias
|
BMD|CMD3B|DXS142|DXS164|DXS206|DXS230|DXS239|DXS268|DXS269|DXS270|DXS272|MRX85 |
|
Transcripts
|
ENSG00000198947
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Cancer-related genes, Disease related genes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Transporters |
|
Uniport_ID
|
P11532
|
|
HGNC ID
|
HGNC:2928
|
|
OMIM ID
|
300377 |
|
Summary
|
This gene spans a genomic range of greater than 2 Mb and encodes a large protein containing an N-terminal actin-binding domain and multiple spectrin repeats. The encoded protein forms a component of the dystrophin-glycoprotein complex (DGC), which bridges the inner cytoskeleton and the extracellular matrix. Deletions, duplications, and point mutations at this gene locus may cause Duchenne muscular dystrophy (DMD), Becker muscular dystrophy (BMD), or cardiomyopathy. Alternative promoter usage and alternative splicing result in numerous distinct transcript variants and protein isoforms for this gene. [provided by RefSeq, Dec 2016] |
Target gene [DMD] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1006936 |
chrX |
11 |
15 |
6 |
32 |
View |
| 1007743 |
chrX |
0 |
1 |
23 |
24 |
View |
| 1012368 |
chrX |
3 |
1 |
3 |
7 |
View |
| 1015725 |
chrX |
0 |
2 |
0 |
2 |
View |
| 1015726 |
chrX |
0 |
2 |
0 |
2 |
View |
| 1019752 |
chrX |
0 |
0 |
0 |
0 |
View |
| 1019753 |
chrX |
0 |
0 |
0 |
0 |
View |
| 1019764 |
chrX |
0 |
0 |
0 |
0 |
View |
| 1019765 |
chrX |
0 |
0 |
0 |
0 |
View |
| 1024658 |
chrX |
0 |
0 |
0 |
0 |
View |
Target gene [DMD] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - DMD peak across samples
|
Peak Plot
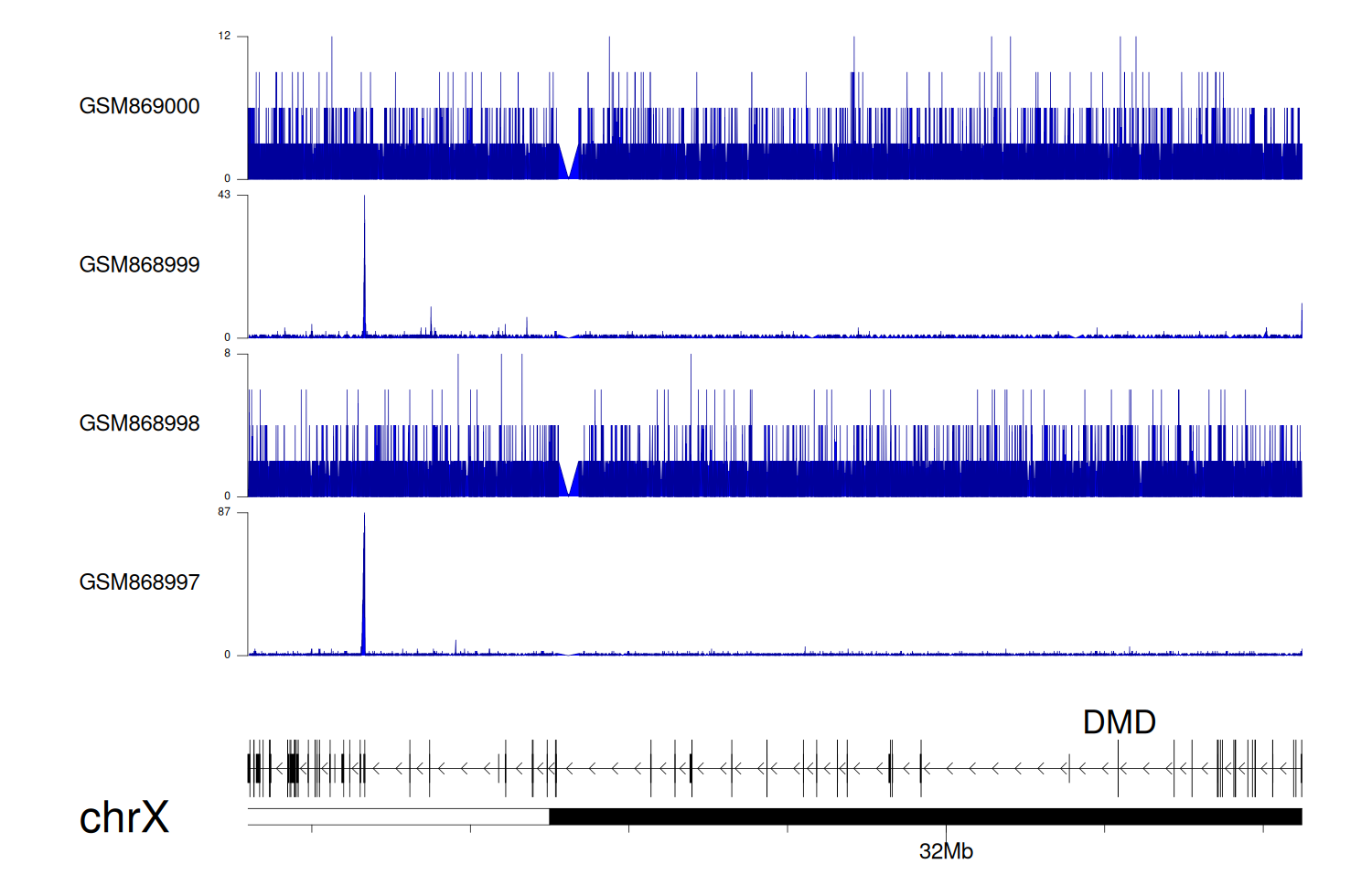
|
> Dataset: GSE68402 - DMD peak across samples
|
Peak Plot
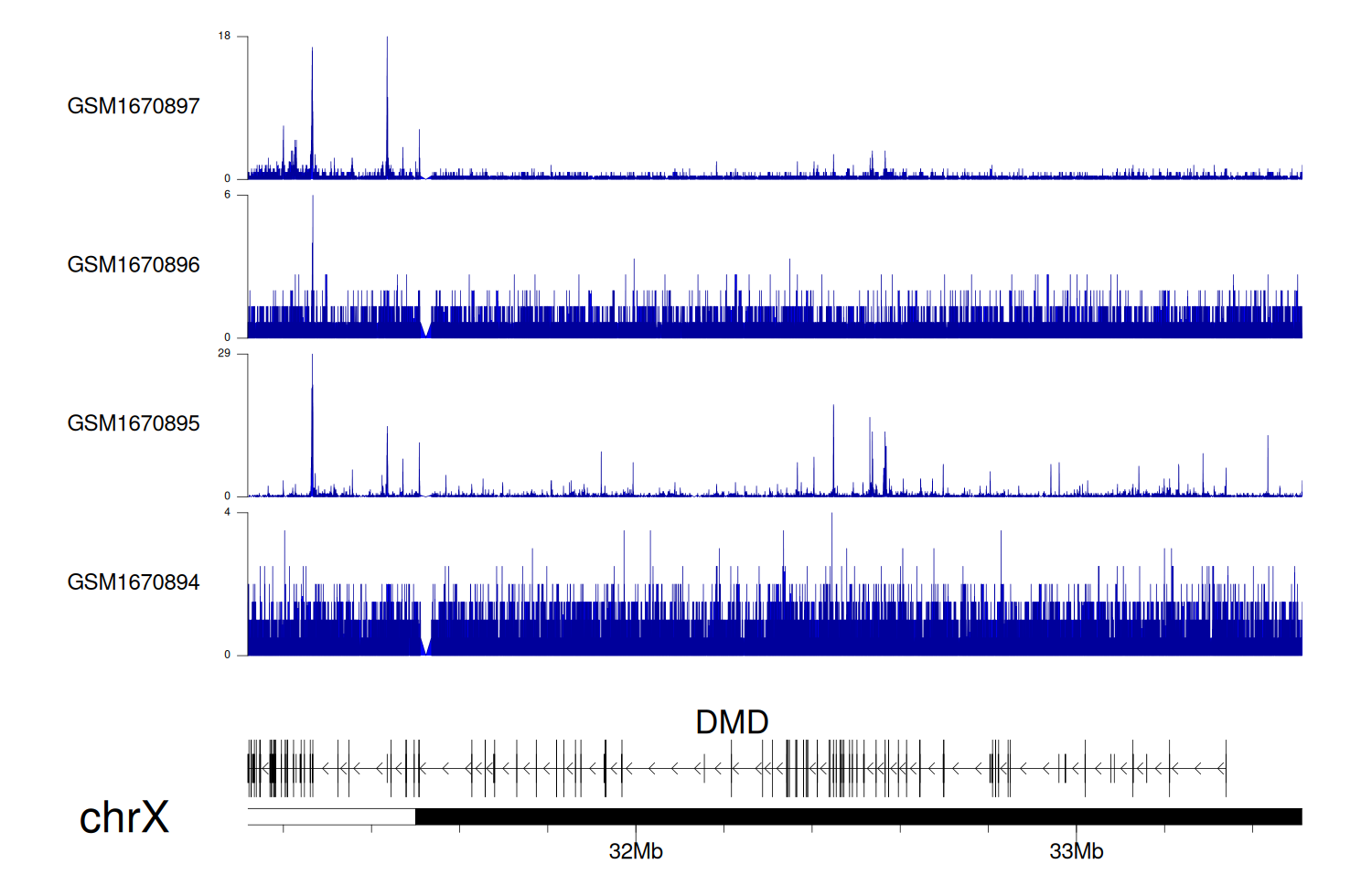
|
> Dataset: GSE270130 - DMD peak across samples
|
Peak Plot
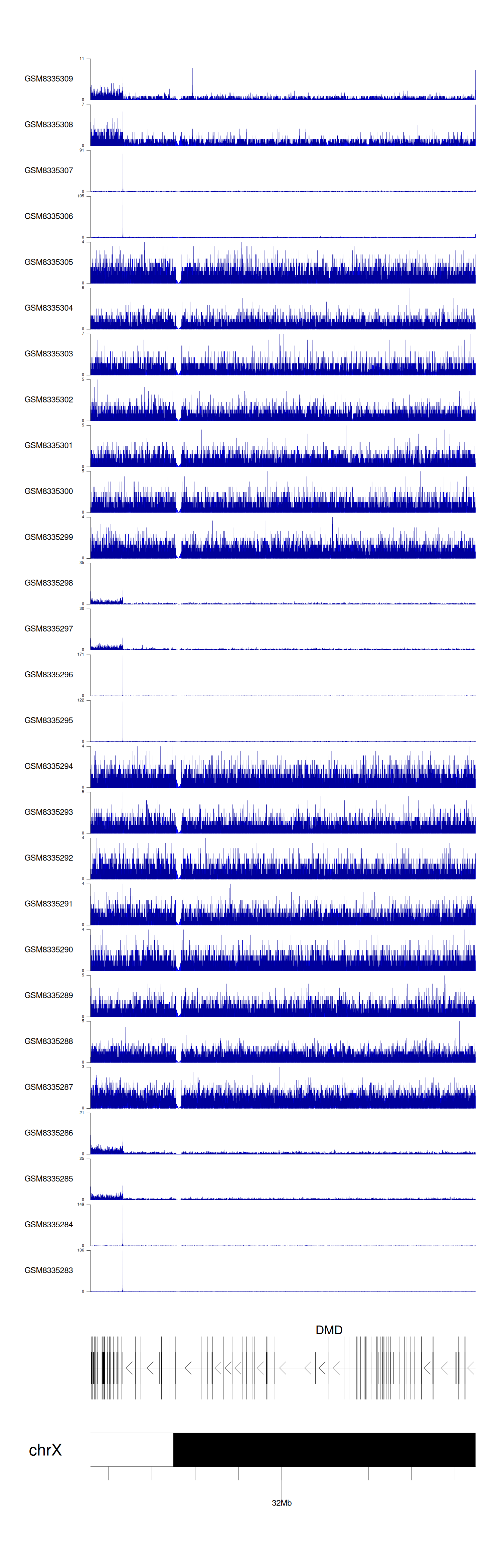
|
> Dataset: GSE100400 - DMD peak across samples
|
Peak Plot
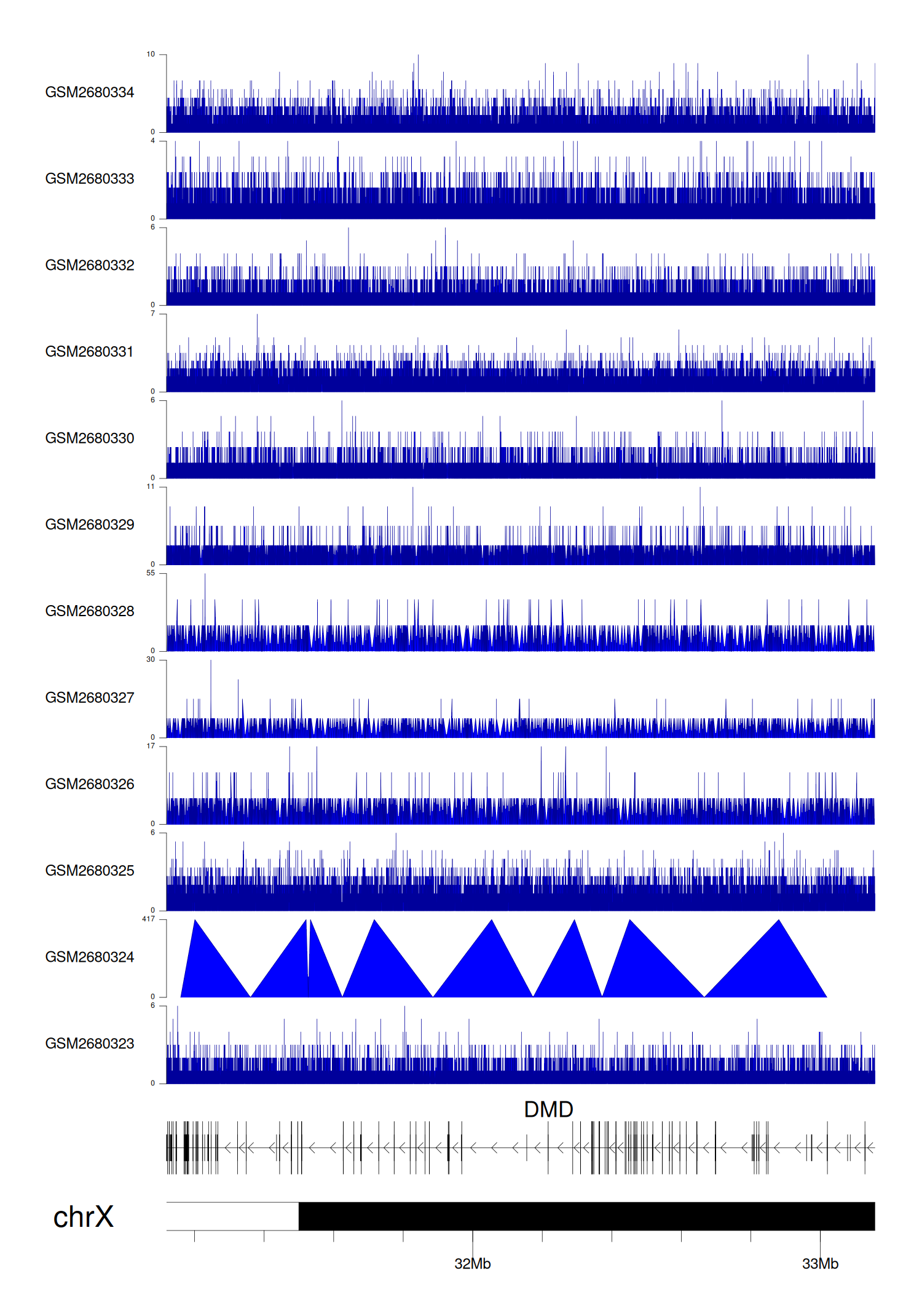
|
> Dataset: GSE131257 - DMD peak across samples
|
Peak Plot
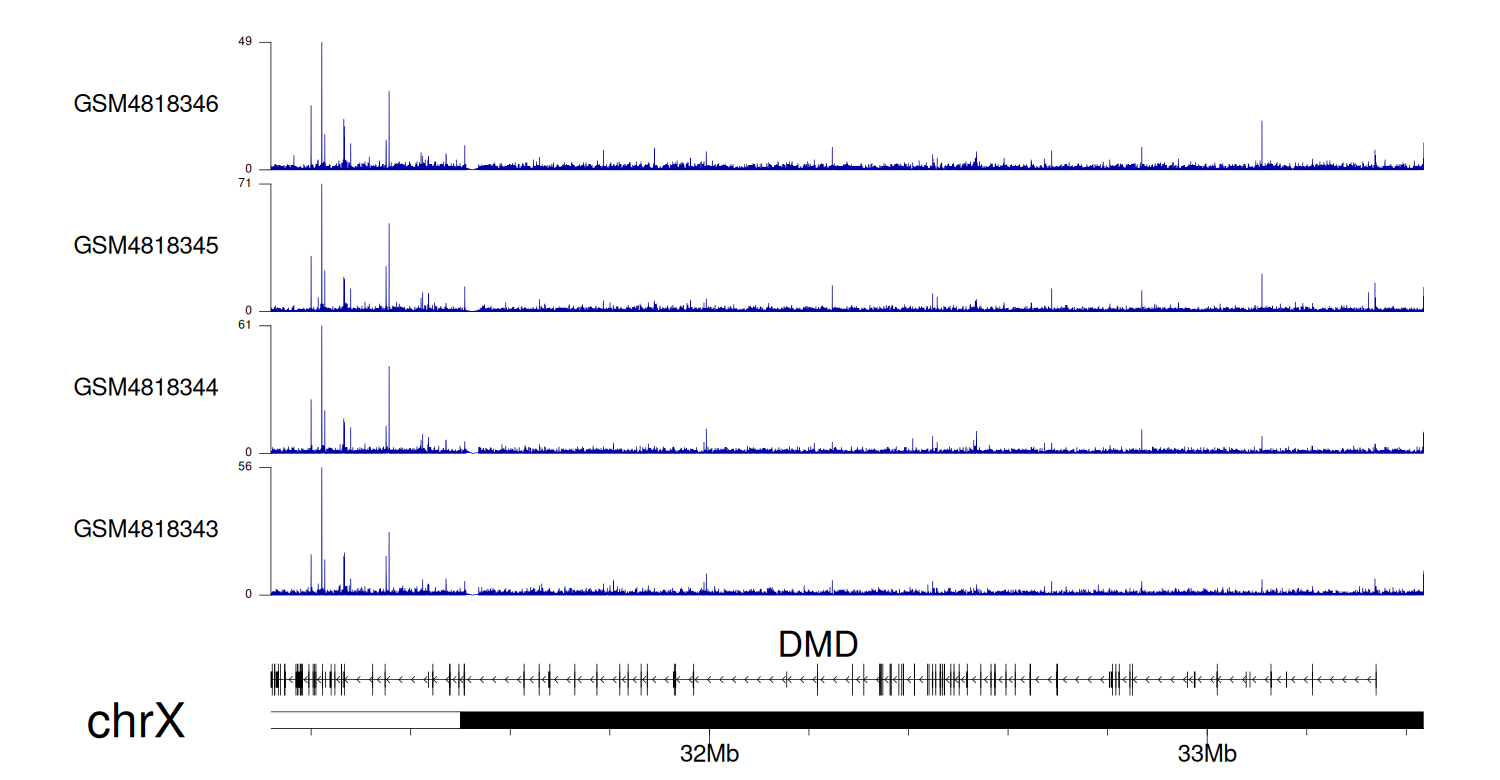
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information