Gene Information
|
Gene Name
|
ENPP1 |
|
Gene ID
|
5167
|
|
Gene Full Name
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
|
Gene Alias
|
ARHR2|COLED|M6S1|NPP1|NPPS|PC-1|PCA1|PDNP1 |
|
Transcripts
|
ENSG00000197594
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Secreted - unknown location; 
|
|
Membrane Info
|
CD markers, Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Potential drug targets, Predicted membrane proteins, Predicted secreted proteins |
|
Uniport_ID
|
P22413
|
|
HGNC ID
|
HGNC:3356
|
|
OMIM ID
|
173335 |
|
Summary
|
This gene is a member of the ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family. The encoded protein is a type II transmembrane glycoprotein comprising two identical disulfide-bonded subunits. This protein has broad specificity and cleaves a variety of substrates, including phosphodiester bonds of nucleotides and nucleotide sugars and pyrophosphate bonds of nucleotides and nucleotide sugars. This protein may function to hydrolyze nucleoside 5' triphosphates to their corresponding monophosphates and may also hydrolyze diadenosine polyphosphates. Mutations in this gene have been associated with 'idiopathic' infantile arterial calcification, ossification of the posterior longitudinal ligament of the spine (OPLL), and insulin resistance. [provided by RefSeq, Jul 2008] |
Target gene [ENPP1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1012384 |
chr6 |
4 |
3 |
0 |
7 |
View |
Target gene [ENPP1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - ENPP1 peak across samples
|
Peak Plot
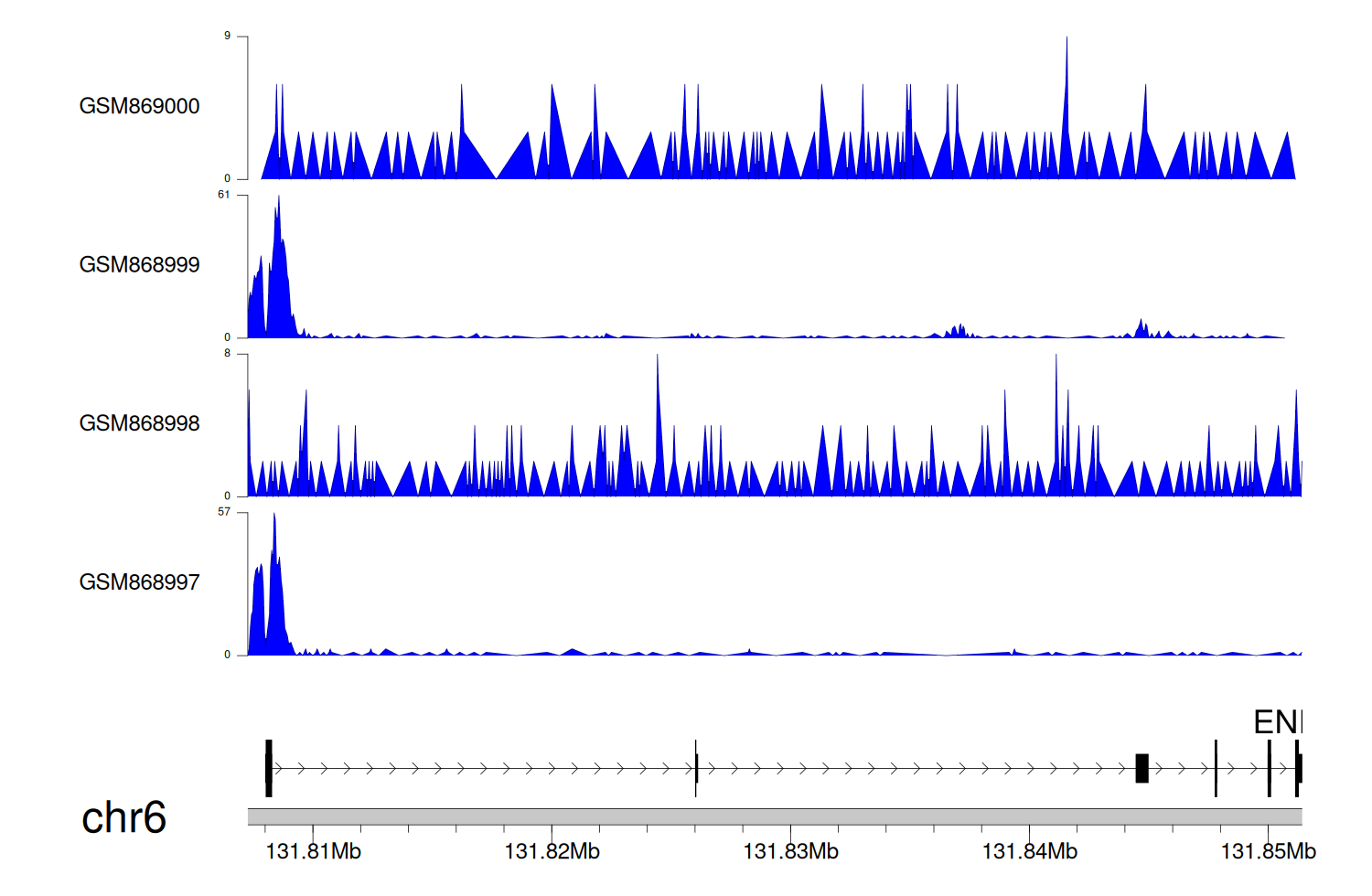
|
> Dataset: GSE68402 - ENPP1 peak across samples
|
Peak Plot
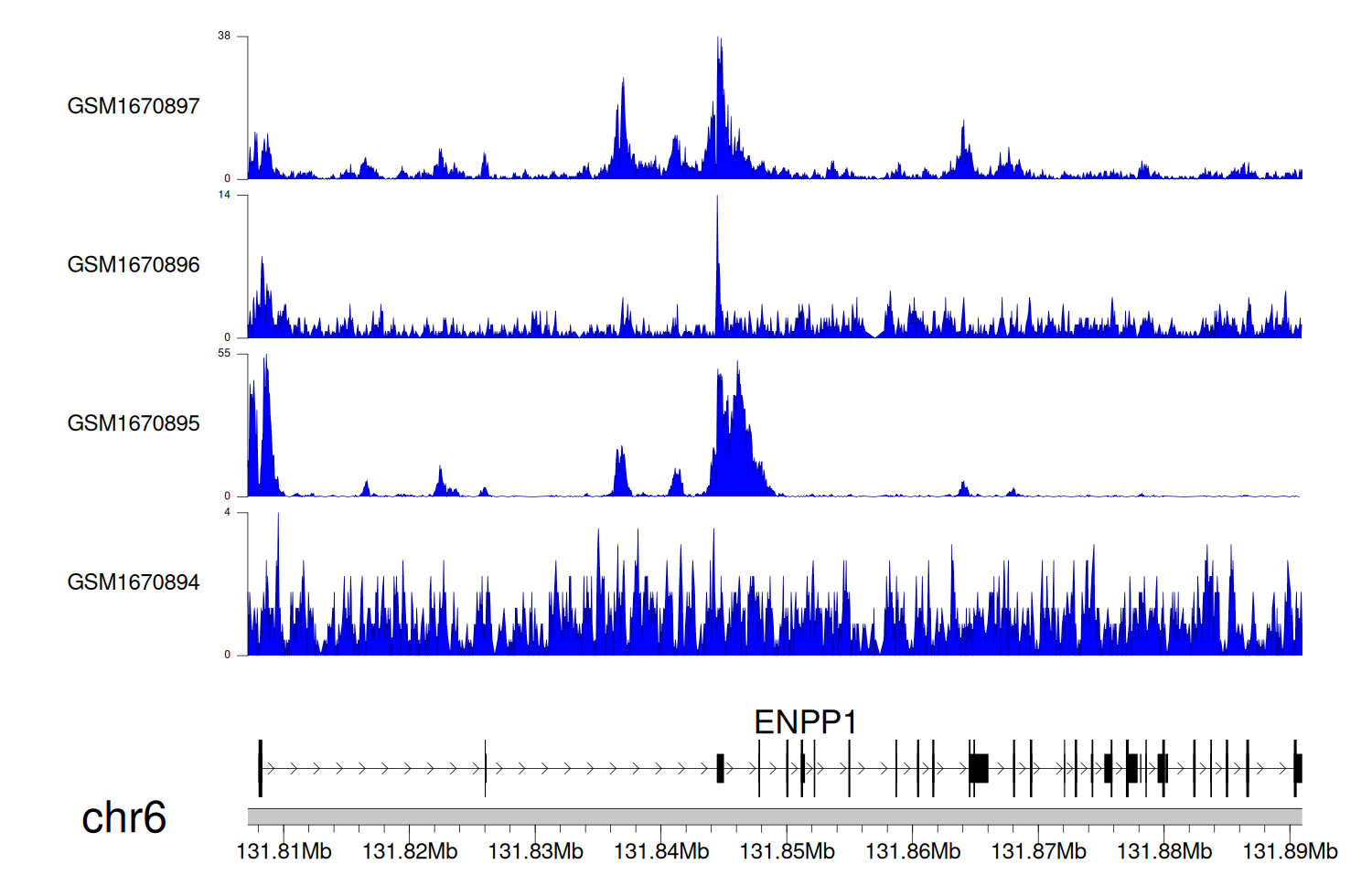
|
> Dataset: GSE270130 - ENPP1 peak across samples
|
Peak Plot
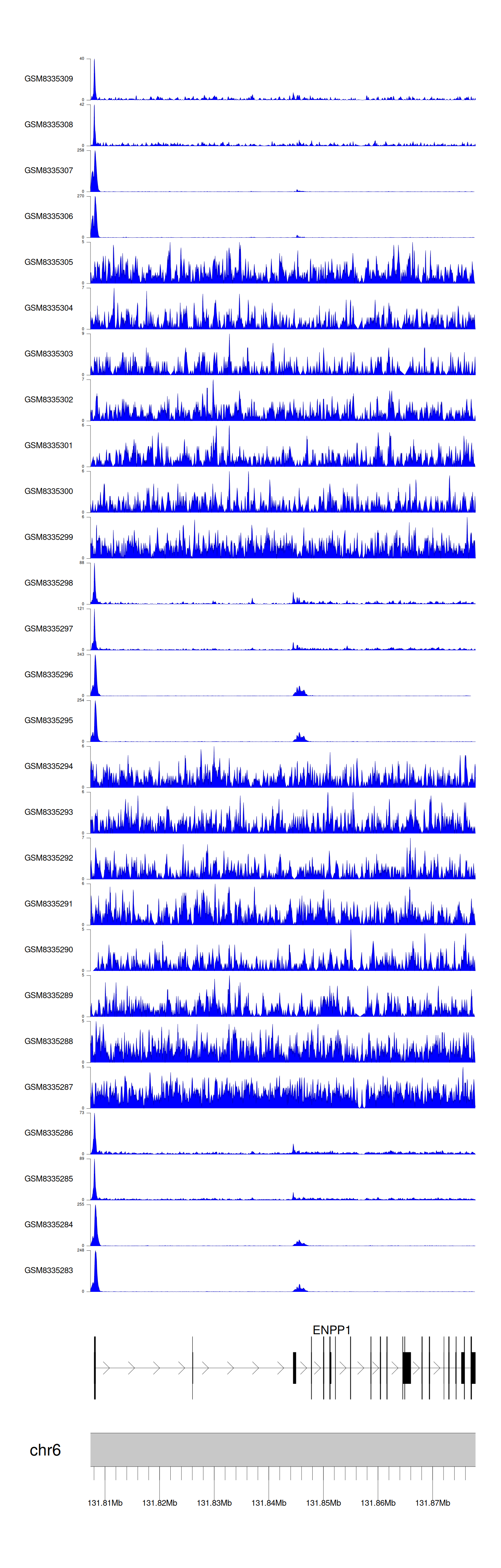
|
> Dataset: GSE131257 - ENPP1 peak across samples
|
Peak Plot
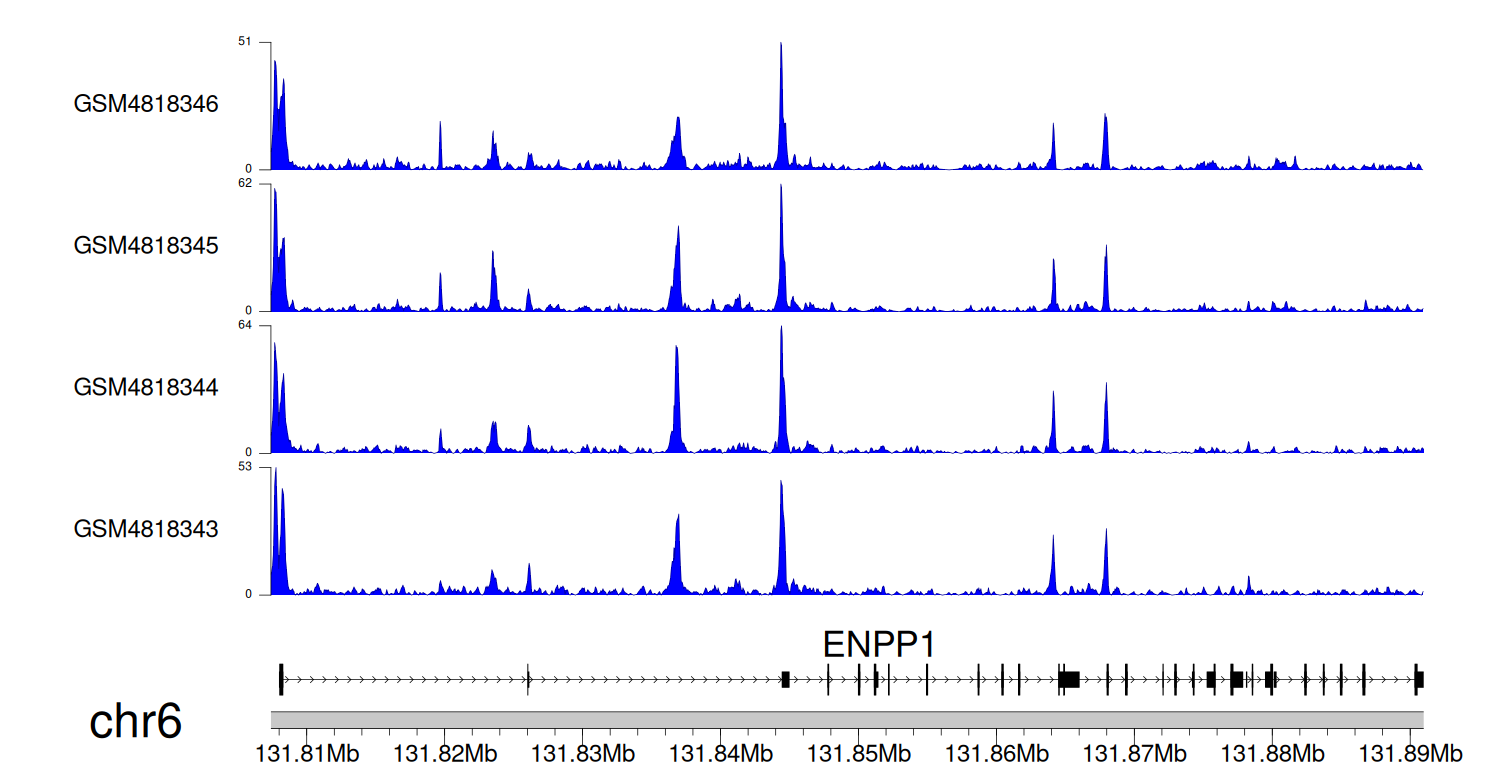
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information