Gene Information
|
Gene Name
|
KAZN |
|
Gene ID
|
23254
|
|
Gene Full Name
|
kazrin, periplakin interacting protein |
|
Gene Alias
|
C1orf196|KAZ |
|
Transcripts
|
ENSG00000189337
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Nuclear speckles, Cytosol; 
|
|
Membrane Info
|
Predicted intracellular proteins |
|
Uniport_ID
|
Q674X7
|
|
HGNC ID
|
HGNC:29173
|
|
OMIM ID
|
618301 |
|
Summary
|
This gene encodes a protein that plays a role in desmosome assembly, cell adhesion, cytoskeletal organization, and epidermal differentiation. This protein co-localizes with desmoplakin and the cytolinker protein periplakin. In general, this protein localizes to the nucleus, desmosomes, cell membrane, and cortical actin-based structures. Some isoforms of this protein also associate with microtubules. Alternative splicing results in multiple transcript variants encoding distinct isoforms. Additional splice variants have been described but their biological validity has not been verified. [provided by RefSeq, Aug 2011] |
Target gene [KAZN] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1001602 |
chr1 |
551 |
11 |
24 |
586 |
View |
| 1006326 |
chr1 |
4 |
21 |
15 |
40 |
View |
| 1007884 |
chr1 |
23 |
0 |
0 |
23 |
View |
| 1010706 |
chr1 |
5 |
6 |
0 |
11 |
View |
| 1016393 |
chr1 |
0 |
1 |
0 |
1 |
View |
| 1017223 |
chr1 |
0 |
1 |
0 |
1 |
View |
Target gene [KAZN] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - KAZN expression across samples
|
Volcano Plot
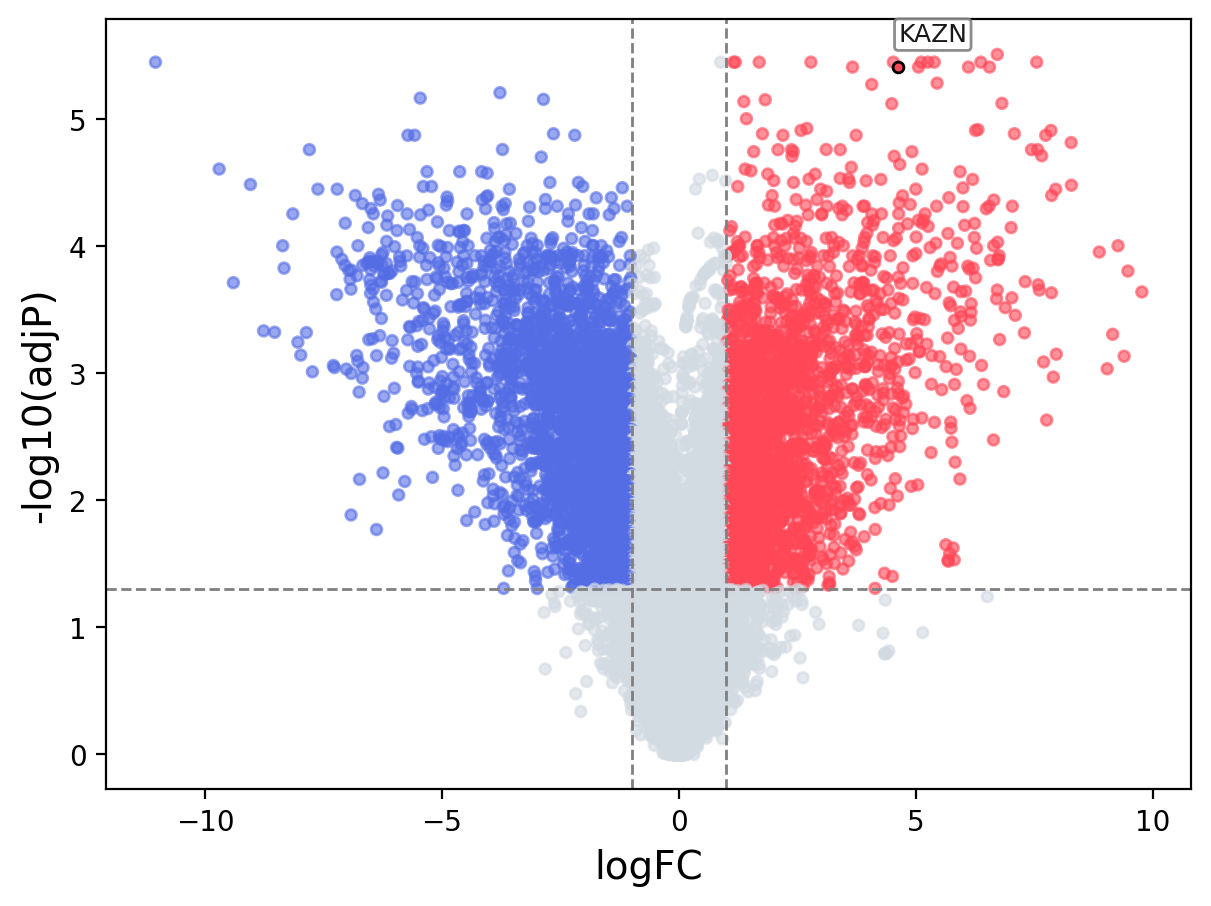
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information