Gene Information
|
Gene Name
|
KCNJ3 |
|
Gene ID
|
3760
|
|
Gene Full Name
|
potassium inwardly rectifying channel subfamily J member 3 |
|
Gene Alias
|
GIRK1|KGA|KIR3.1 |
|
Transcripts
|
ENSG00000162989
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Mitochondria;Plasma membrane; 
|
|
Membrane Info
|
FDA approved drug targets, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins, Transporters, Voltage-gated ion channels |
|
Uniport_ID
|
P48549
|
|
HGNC ID
|
HGNC:6264
|
|
OMIM ID
|
601534 |
|
Summary
|
Potassium channels are present in most mammalian cells, where they participate in a wide range of physiologic responses. The protein encoded by this gene is an integral membrane protein and inward-rectifier type potassium channel. The encoded protein, which has a greater tendency to allow potassium to flow into a cell rather than out of a cell, is controlled by G-proteins and plays an important role in regulating heartbeat. It associates with three other G-protein-activated potassium channels to form a heteromultimeric pore-forming complex that also couples to neurotransmitter receptors in the brain and whereby channel activation can inhibit action potential firing by hyperpolarizing the plasma membrane. These multimeric G-protein-gated inwardly-rectifying potassium (GIRK) channels may play a role in the pathophysiology of epilepsy, addiction, Down's syndrome, ataxia, and Parkinson's disease. Alternative splicing results in multiple transcript variants encoding distinct proteins. [provided by RefSeq, May 2012] |
Target gene [KCNJ3] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1002323 |
chr2 |
32 |
237 |
24 |
293 |
View |
| 1002335 |
chr2 |
33 |
233 |
24 |
290 |
View |
| 1020557 |
chr2 |
41 |
1 |
3 |
45 |
View |
| 1041281 |
chr2 |
0 |
0 |
0 |
0 |
View |
Target gene [KCNJ3] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
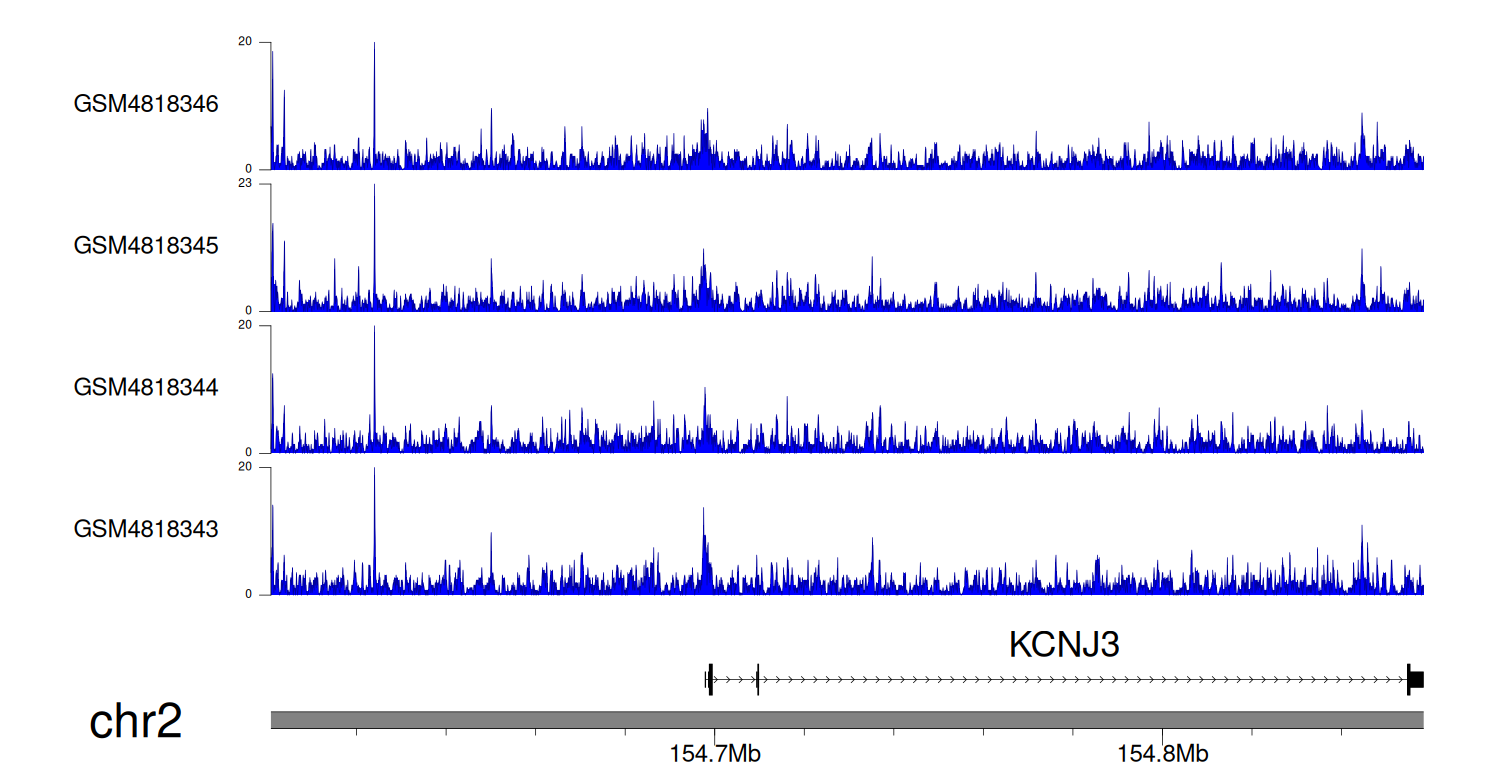
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - KCNJ3 peak across samples
|
Peak Plot
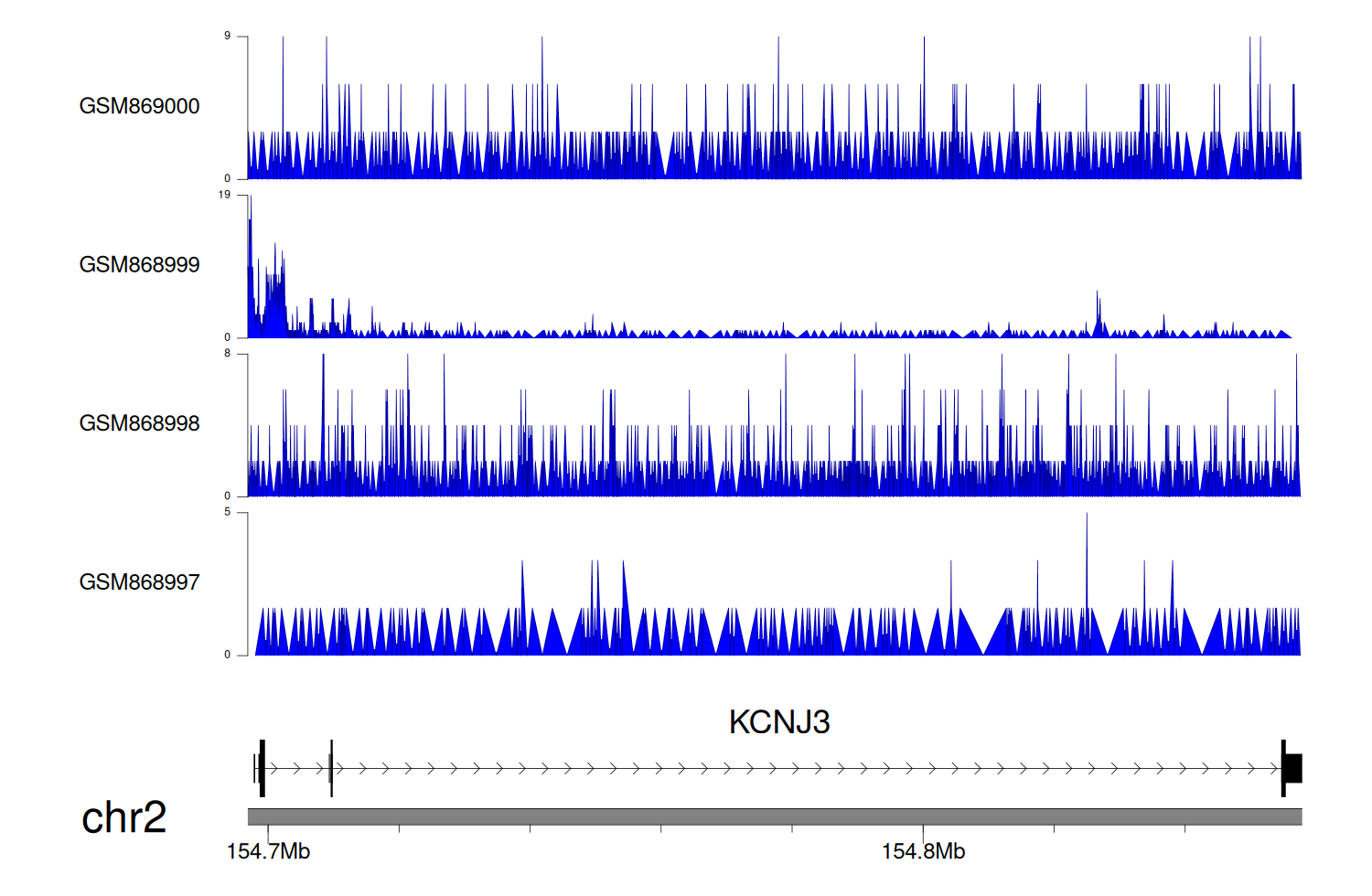
|
> Dataset: GSE68402 - KCNJ3 peak across samples
|
Peak Plot
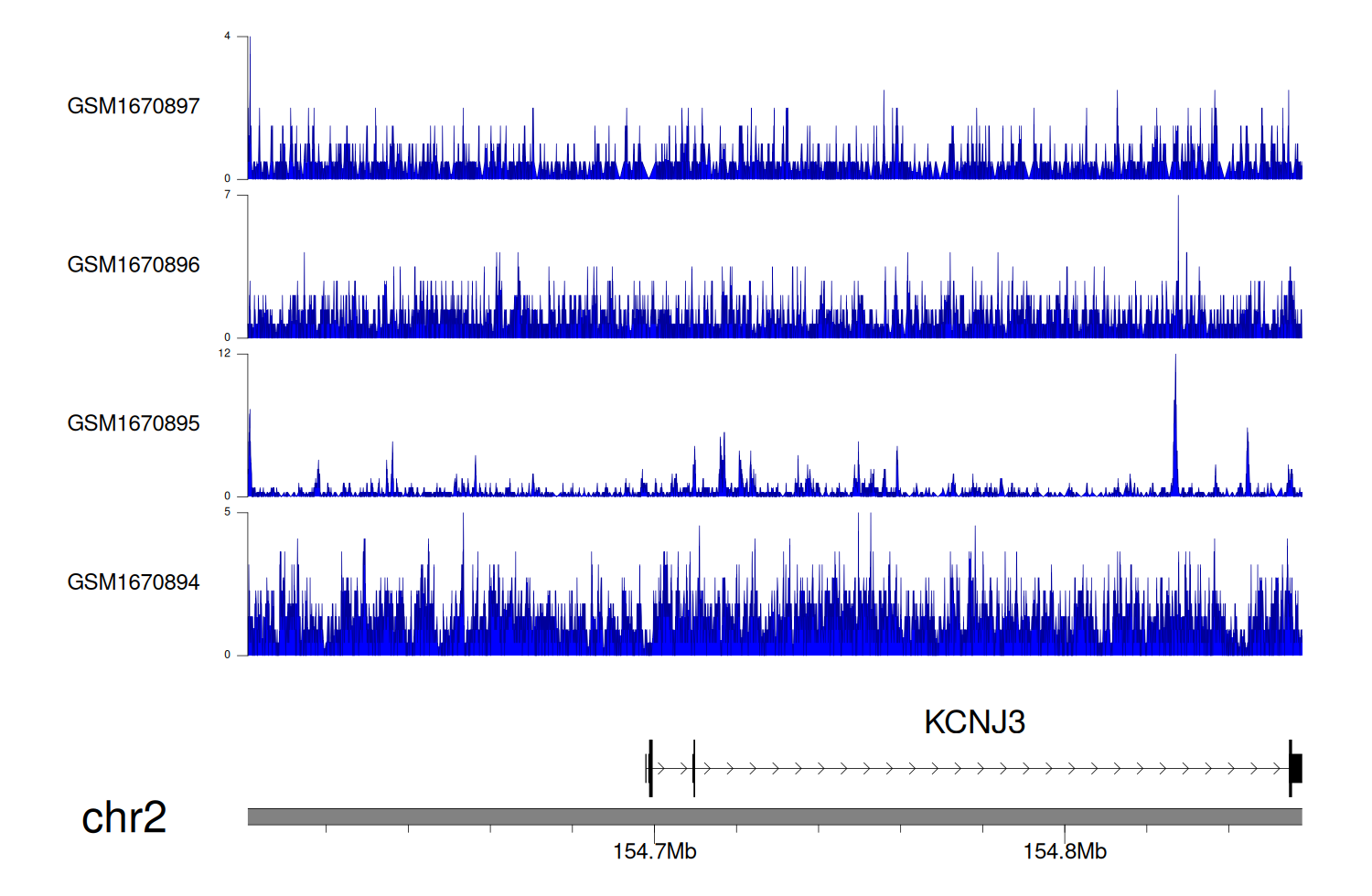
|
> Dataset: GSE270130 - KCNJ3 peak across samples
|
Peak Plot
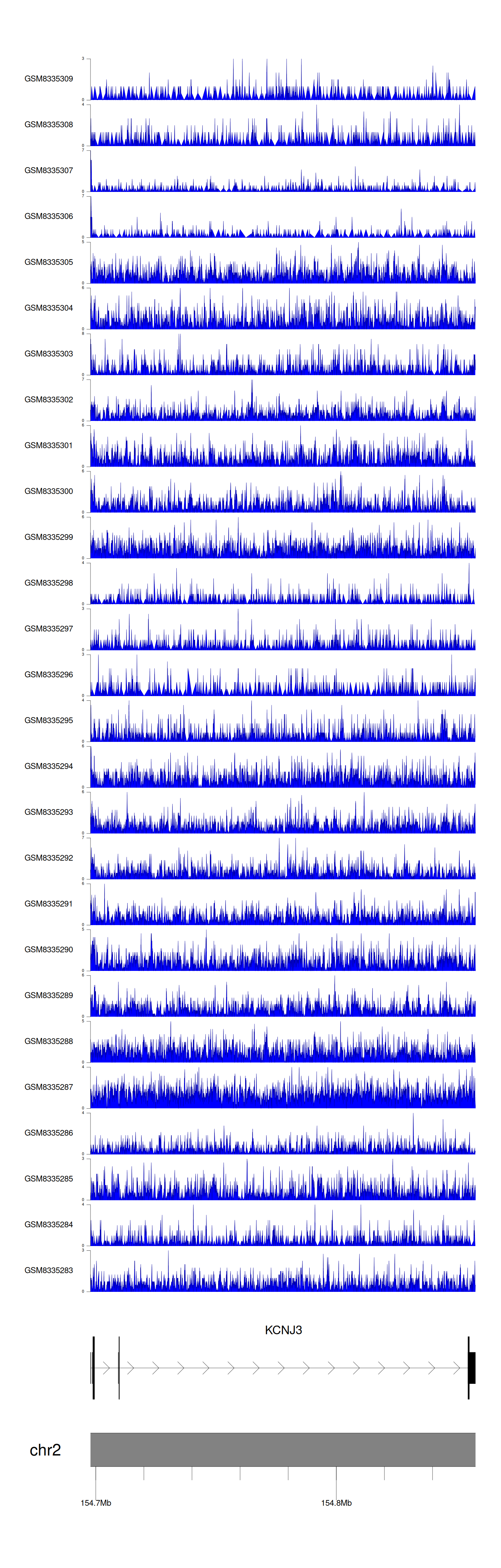
|
> Dataset: GSE100400 - KCNJ3 peak across samples
|
Peak Plot
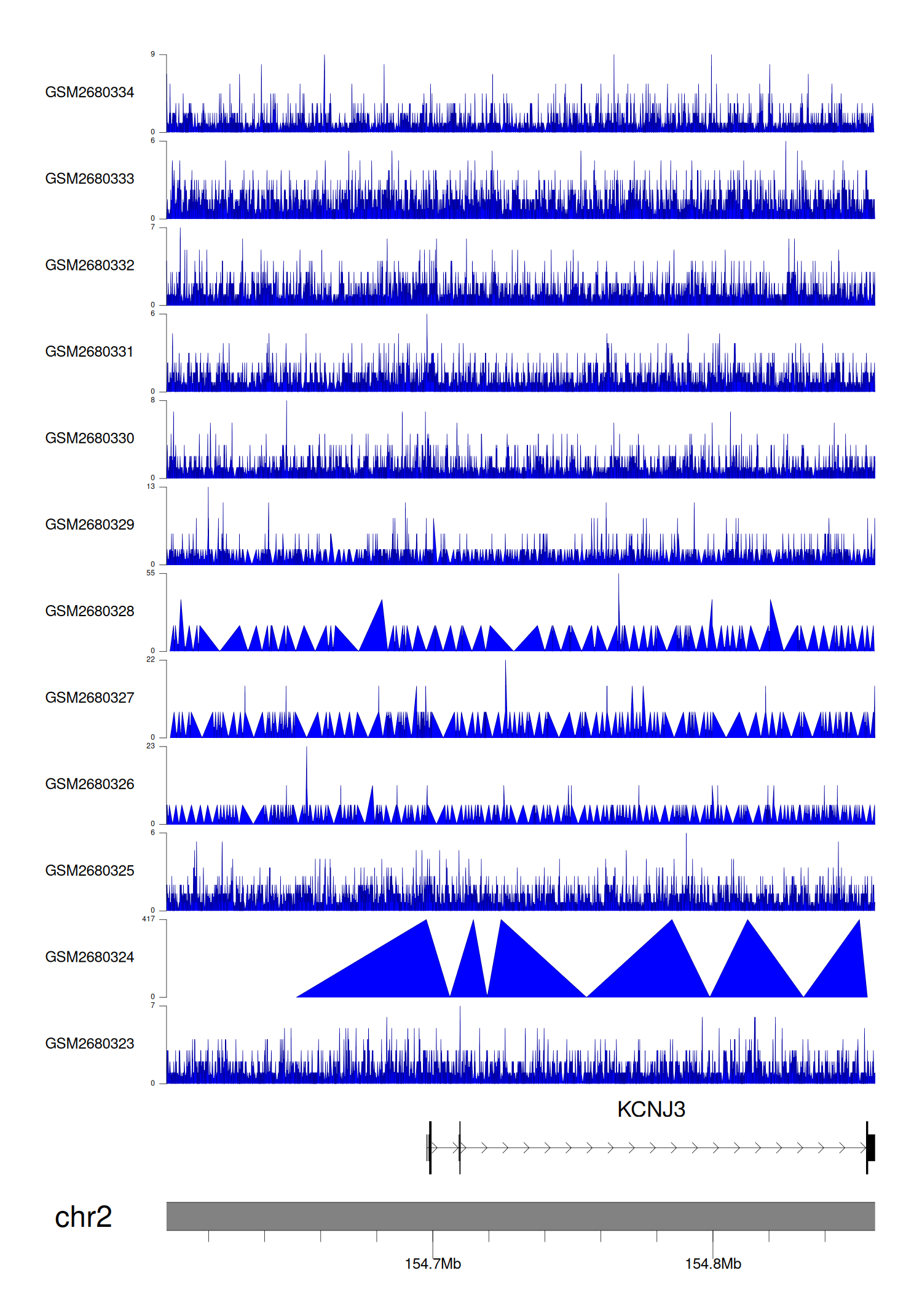
|
> Dataset: GSE131257 - KCNJ3 peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information