Gene Information
|
Gene Name
|
MME |
|
Gene ID
|
4311
|
|
Gene Full Name
|
membrane metalloendopeptidase |
|
Gene Alias
|
CALLA|CD10|CMT2T|NEP|SCA43|SFE |
|
Transcripts
|
ENSG00000196549
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane, Centrosome, Basal body;Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Candidate cardiovascular disease genes, CD markers, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P08473
|
|
HGNC ID
|
HGNC:7154
|
|
OMIM ID
|
120520 |
|
Summary
|
The protein encoded by this gene is a type II transmembrane glycoprotein and a common acute lymphocytic leukemia antigen that is an important cell surface marker in the diagnosis of human acute lymphocytic leukemia (ALL). The encoded protein is present on leukemic cells of pre-B phenotype, which represent 85% of cases of ALL. This protein is not restricted to leukemic cells, however, and is found on a variety of normal tissues. The protein is a neutral endopeptidase that cleaves peptides at the amino side of hydrophobic residues and inactivates several peptide hormones including glucagon, enkephalins, substance P, neurotensin, oxytocin, and bradykinin. [provided by RefSeq, Aug 2017] |
Target gene [MME] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1003089 |
chr3 |
78 |
17 |
72 |
167 |
View |
| 1003115 |
chr3 |
76 |
17 |
71 |
164 |
View |
Target gene [MME] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - MME expression across samples
|
Volcano Plot
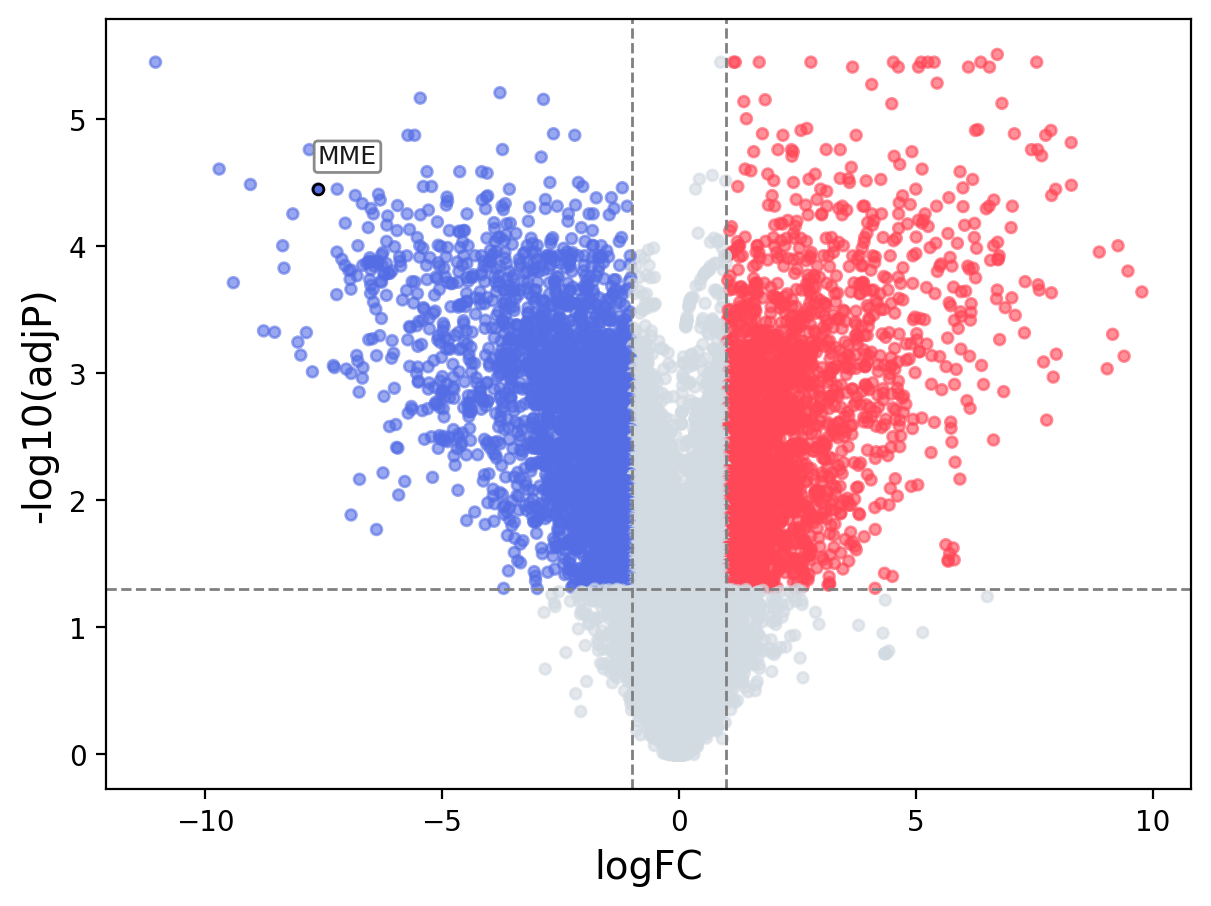
|
Bar Plot
|
> Dataset: GSE94660 - MME expression across samples
|
Volcano Plot
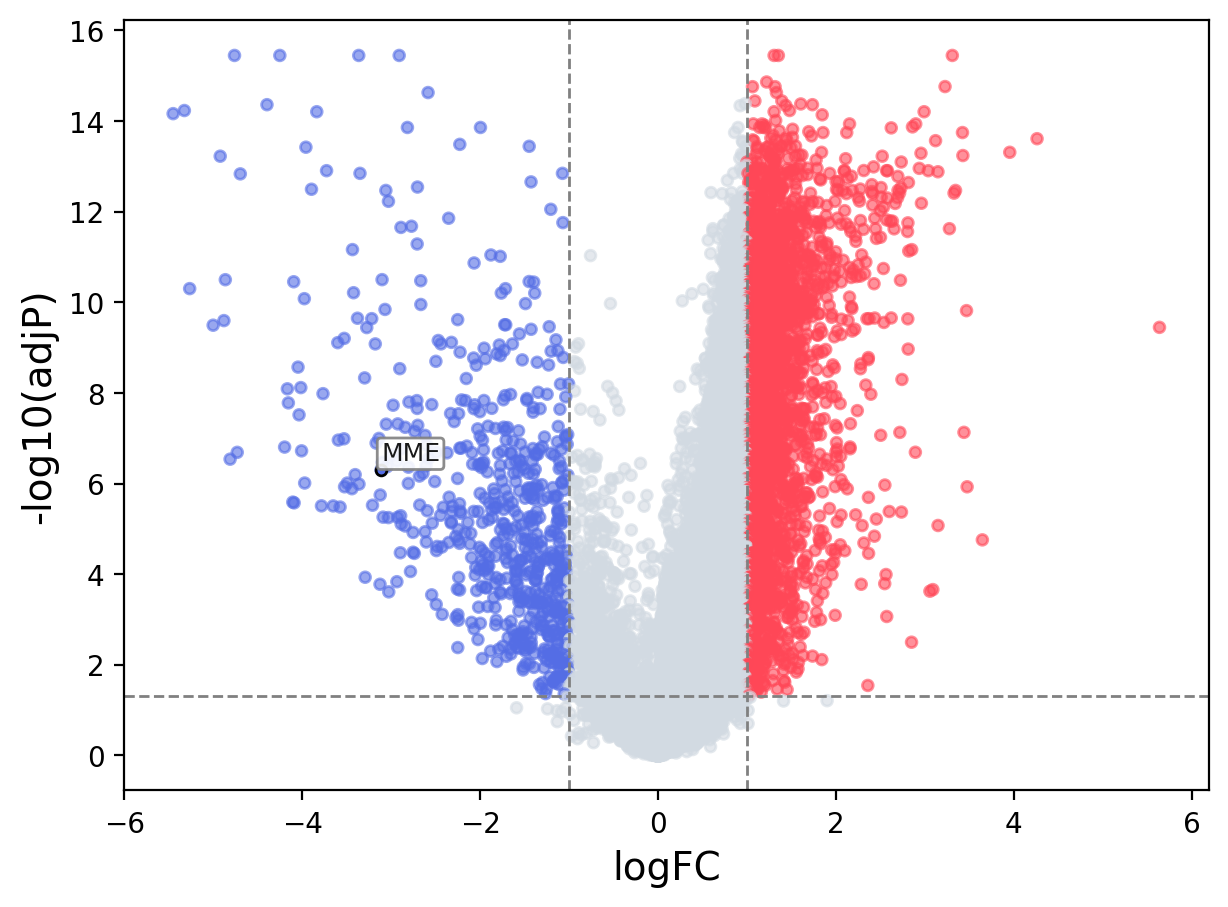
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information