Gene Information
|
Gene Name
|
NTRK3 |
|
Gene ID
|
4916
|
|
Gene Full Name
|
neurotrophic receptor tyrosine kinase 3 |
|
Gene Alias
|
GP145-TrkC|TRKC|gp145(trkC) |
|
Transcripts
|
ENSG00000140538
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoli, Nucleoli rim;Nuclear membrane; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
Q16288
|
|
HGNC ID
|
HGNC:8033
|
|
OMIM ID
|
191316 |
|
Summary
|
This gene encodes a member of the neurotrophic tyrosine receptor kinase (NTRK) family. This kinase is a membrane-bound receptor that, upon neurotrophin binding, phosphorylates itself and members of the MAPK pathway. Signalling through this kinase leads to cell differentiation and may play a role in the development of proprioceptive neurons that sense body position. Mutations in this gene have been associated with medulloblastomas, secretory breast carcinomas and other cancers. Several transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2011] |
Target gene [NTRK3] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1008188 |
chr15 |
13 |
1 |
8 |
22 |
View |
| 1008189 |
chr15 |
15 |
1 |
6 |
22 |
View |
| 1009943 |
chr15 |
13 |
1 |
0 |
14 |
View |
| 1017584 |
chr15 |
0 |
0 |
0 |
0 |
View |
| 1042147 |
chr15 |
5 |
2 |
0 |
7 |
View |
Target gene [NTRK3] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE68402 - NTRK3 peak across samples
|
Peak Plot
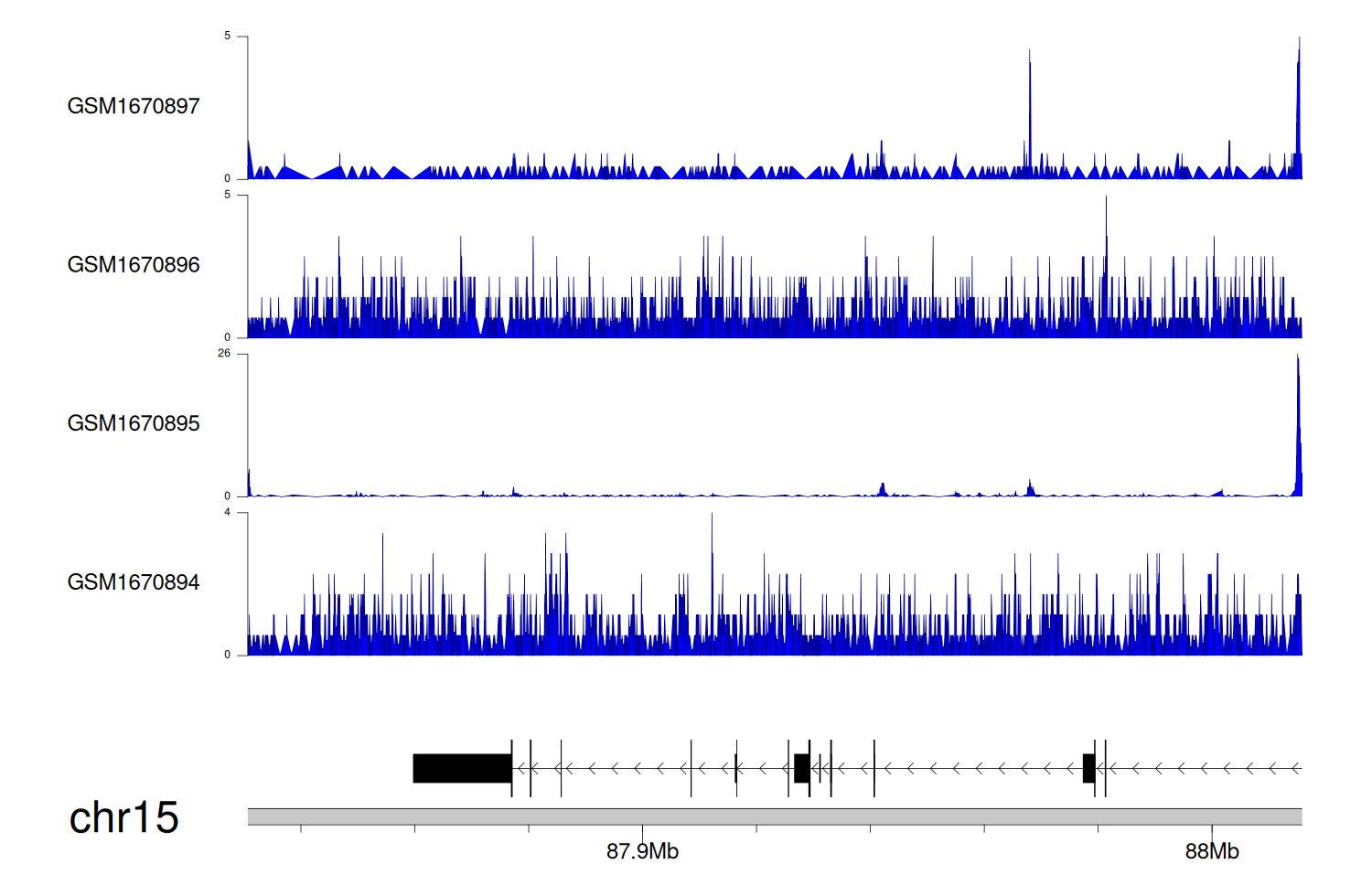
|
> Dataset: GSE270130 - NTRK3 peak across samples
|
Peak Plot
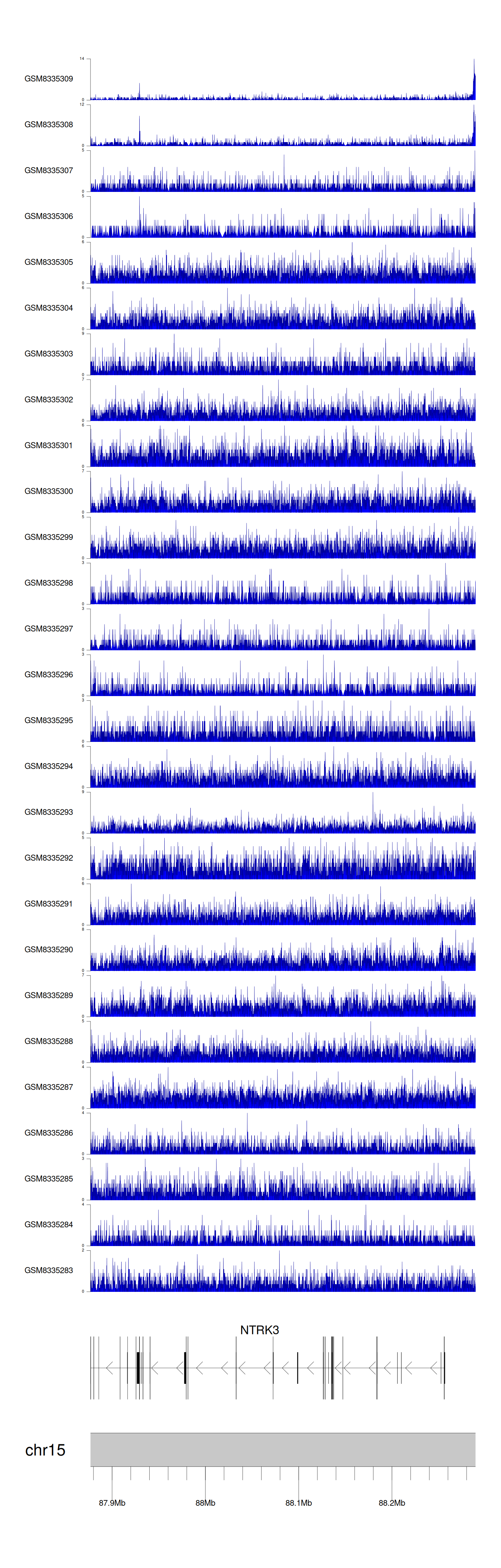
|
> Dataset: GSE100400 - NTRK3 peak across samples
|
Peak Plot

|
> Dataset: GSE131257 - NTRK3 peak across samples
|
Peak Plot
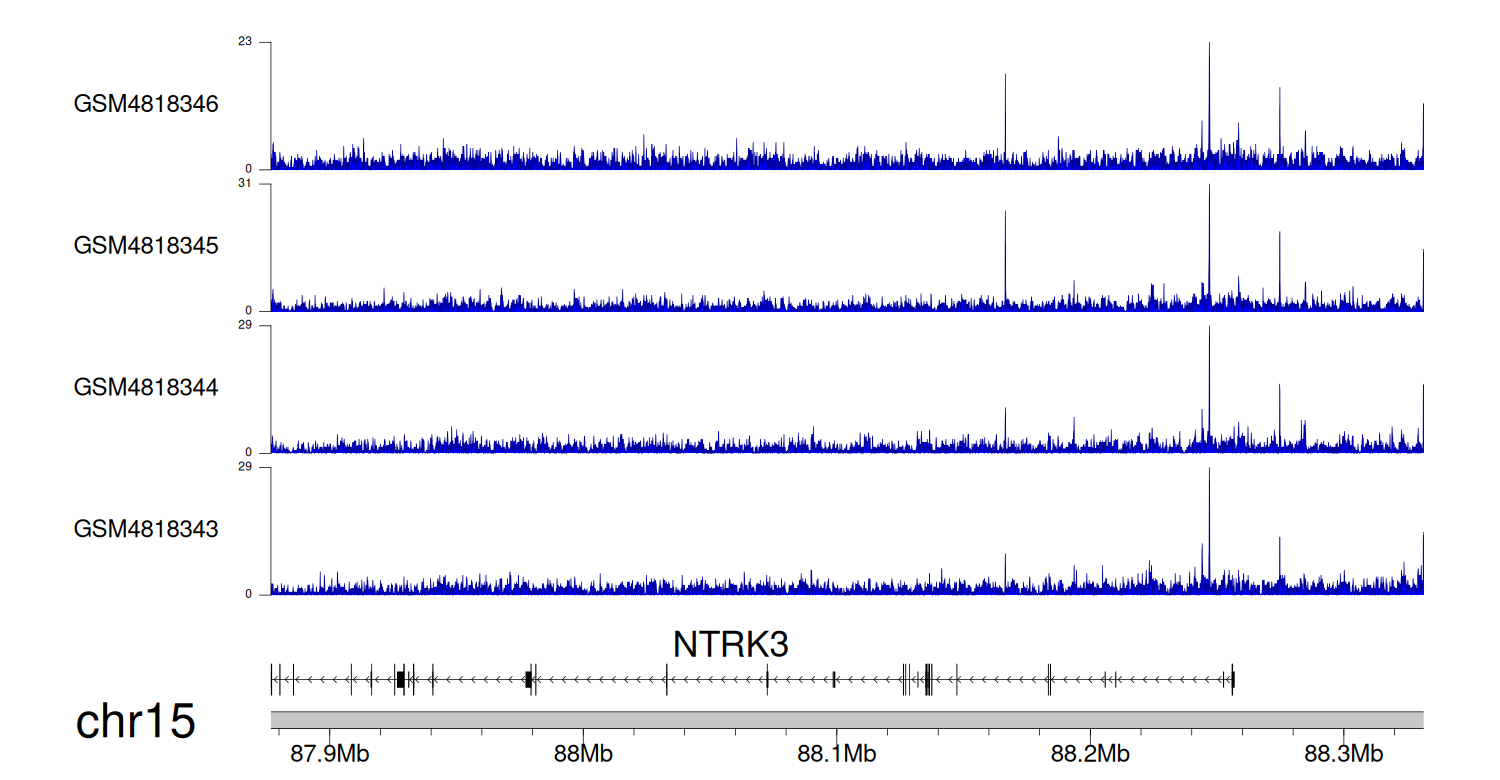
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information