Gene Information
|
Gene Name
|
PATZ1 |
|
Gene ID
|
23598
|
|
Gene Full Name
|
POZ/BTB and AT hook containing zinc finger 1 |
|
Gene Alias
|
MAZR|PATZ|RIAZ|ZBTB19|ZNF278|ZSG|dJ400N23 |
|
Transcripts
|
ENSG00000100105
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Predicted intracellular proteins, Transcription factors |
|
Uniport_ID
|
Q9HBE1
|
|
HGNC ID
|
HGNC:13071
|
|
OMIM ID
|
605165 |
|
Summary
|
The protein encoded by this gene contains an A-T hook DNA binding motif which usually binds to other DNA binding structures to play an important role in chromatin modeling and transcription regulation. Its Poz domain is thought to function as a site for protein-protein interaction and is required for transcriptional repression, and the zinc-fingers comprise the DNA binding domain. Since the encoded protein has typical features of a transcription factor, it is postulated to be a repressor of gene expression. In small round cell sarcoma, this gene is fused to EWS by a small inversion of 22q, then the hybrid is thought to be translocated (t(1;22)(p36.1;q12). The rearrangement of chromosome 22 involves intron 8 of EWS and exon 1 of this gene creating a chimeric sequence containing the transactivation domain of EWS fused to zinc finger domain of this protein. This is a distinct example of an intra-chromosomal rearrangement of chromosome 22. Four alternatively spliced transcript variants are described for this gene. [provided by RefSeq, Jul 2008] |
Target gene [PATZ1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1007176 |
chr22 |
22 |
7 |
0 |
29 |
View |
| 1008470 |
chr22 |
14 |
6 |
0 |
20 |
View |
| 1023496 |
chr22 |
22 |
7 |
0 |
29 |
View |
| 1023497 |
chr22 |
14 |
6 |
0 |
20 |
View |
Target gene [PATZ1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - PATZ1 expression across samples
|
Volcano Plot
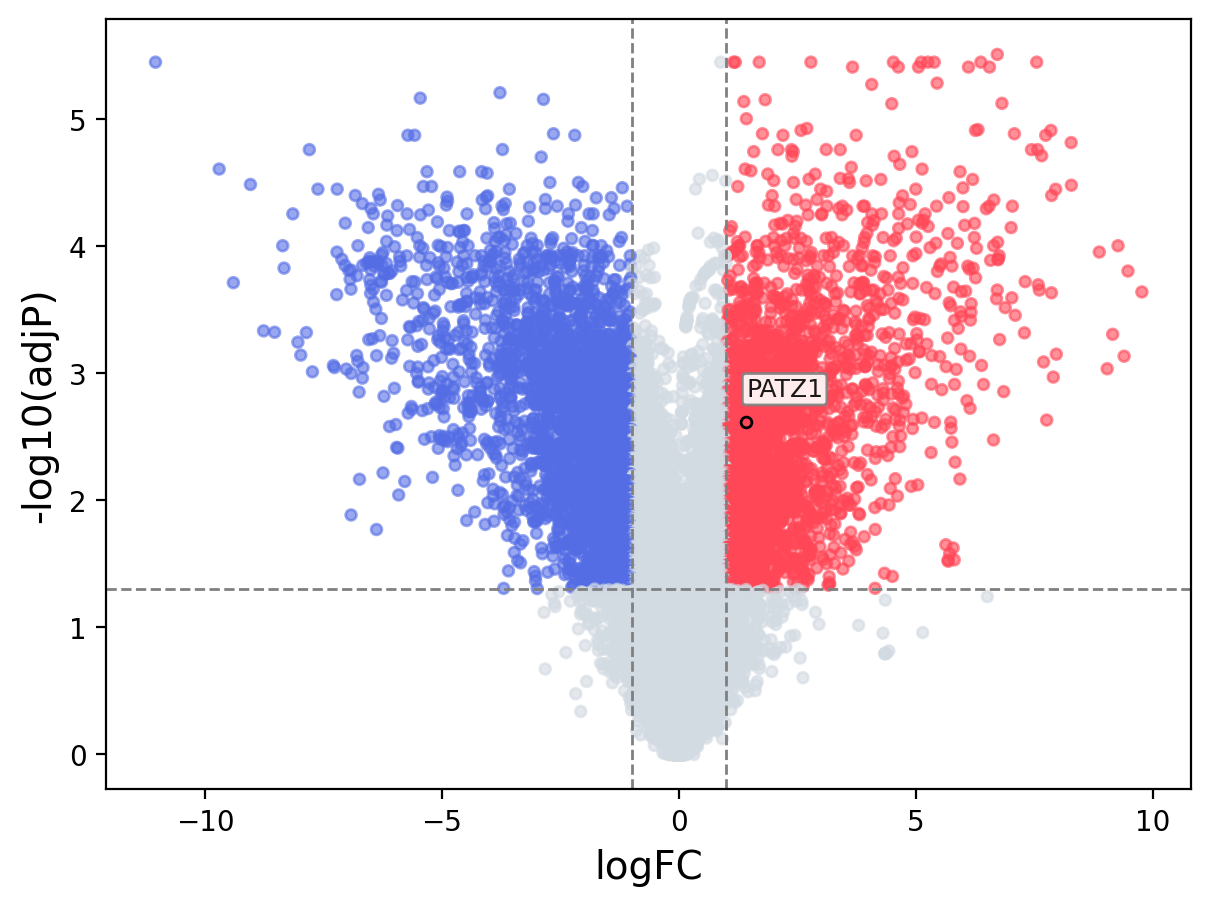
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information