Gene Information
|
Gene Name
|
PCDH15 |
|
Gene ID
|
65217
|
|
Gene Full Name
|
protocadherin related 15 |
|
Gene Alias
|
CDHR15|DFNB23|USH1F |
|
Transcripts
|
ENSG00000150275
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles, Plasma membrane;Nucleoplasm, Golgi apparatus, Focal adhesion sites;Intracellular and membrane; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
Q96QU1
|
|
HGNC ID
|
HGNC:14674
|
|
OMIM ID
|
605514 |
|
Summary
|
This gene is a member of the cadherin superfamily. Family members encode integral membrane proteins that mediate calcium-dependent cell-cell adhesion. It plays an essential role in maintenance of normal retinal and cochlear function. Mutations in this gene result in hearing loss and Usher Syndrome Type IF (USH1F). Extensive alternative splicing resulting in multiple isoforms has been observed in the mouse ortholog. Similar alternatively spliced transcripts are inferred to occur in human, and additional variants are likely to occur. [provided by RefSeq, Dec 2008] |
Target gene [PCDH15] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1010255 |
chr10 |
1 |
12 |
0 |
13 |
View |
| 1011467 |
chr10 |
1 |
3 |
5 |
9 |
View |
| 1016449 |
chr10 |
0 |
1 |
0 |
1 |
View |
| 1017450 |
chr10 |
1 |
0 |
0 |
1 |
View |
| 1019787 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1020016 |
chr10 |
21 |
3 |
1 |
25 |
View |
| 1024572 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1041002 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1041003 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1041816 |
chr10 |
1 |
2 |
0 |
3 |
View |
| 1041817 |
chr10 |
1 |
12 |
0 |
13 |
View |
Target gene [PCDH15] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - PCDH15 peak across samples
|
Peak Plot
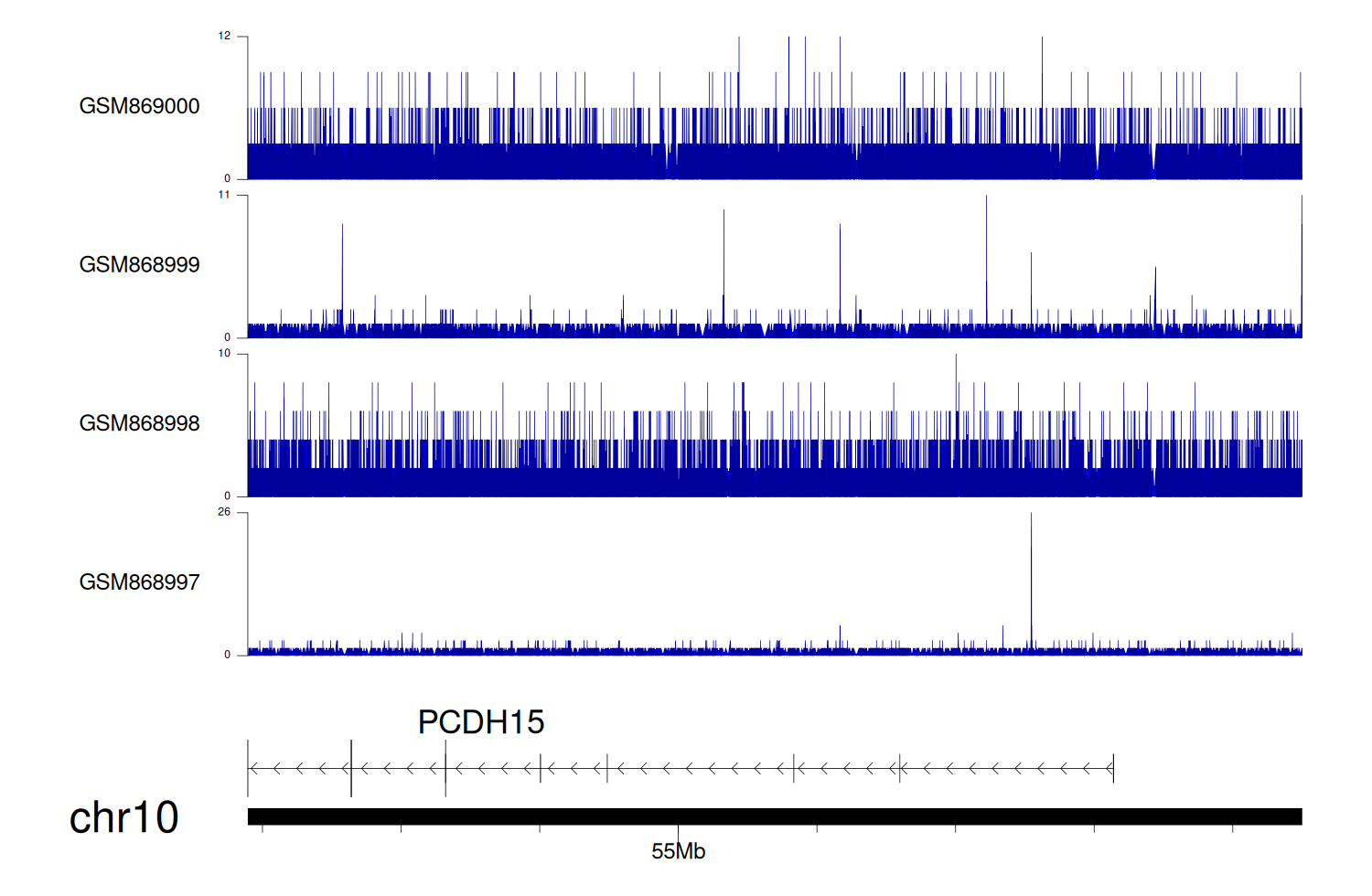
|
> Dataset: GSE68402 - PCDH15 peak across samples
|
Peak Plot
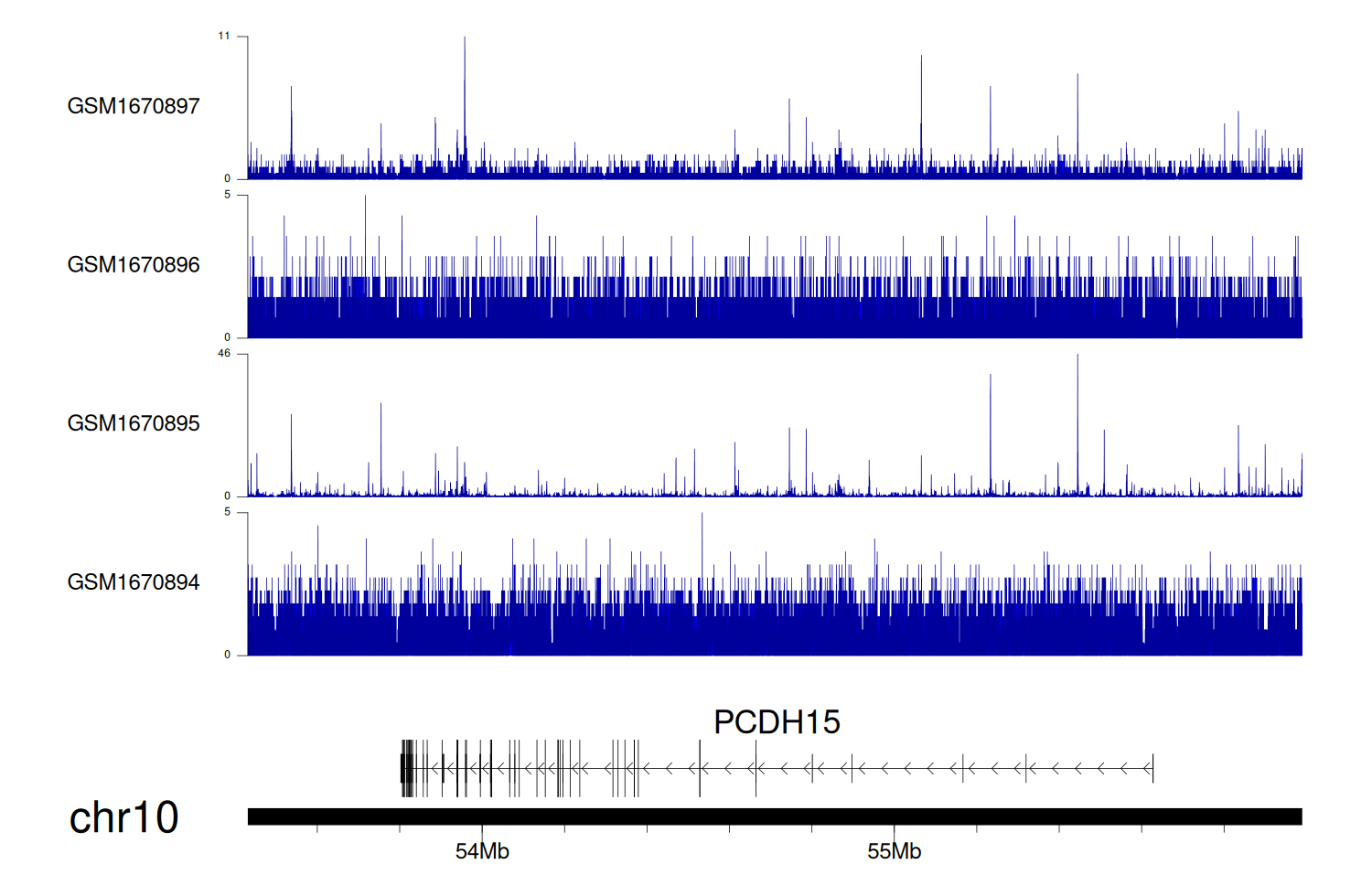
|
> Dataset: GSE270130 - PCDH15 peak across samples
|
Peak Plot
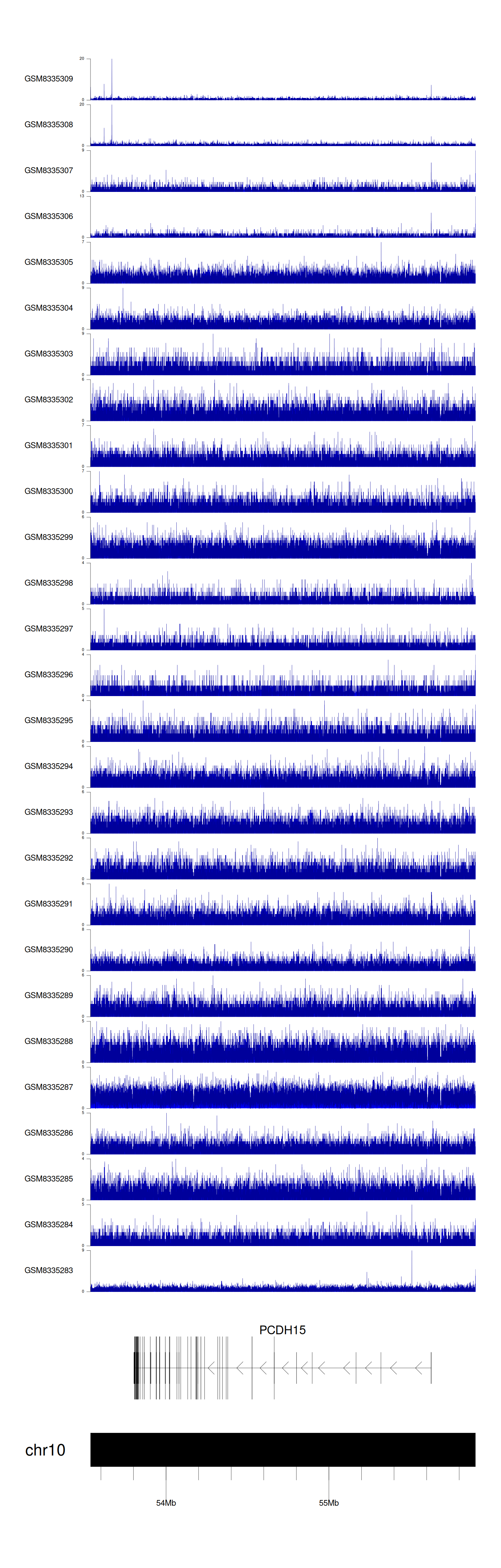
|
> Dataset: GSE100400 - PCDH15 peak across samples
|
Peak Plot
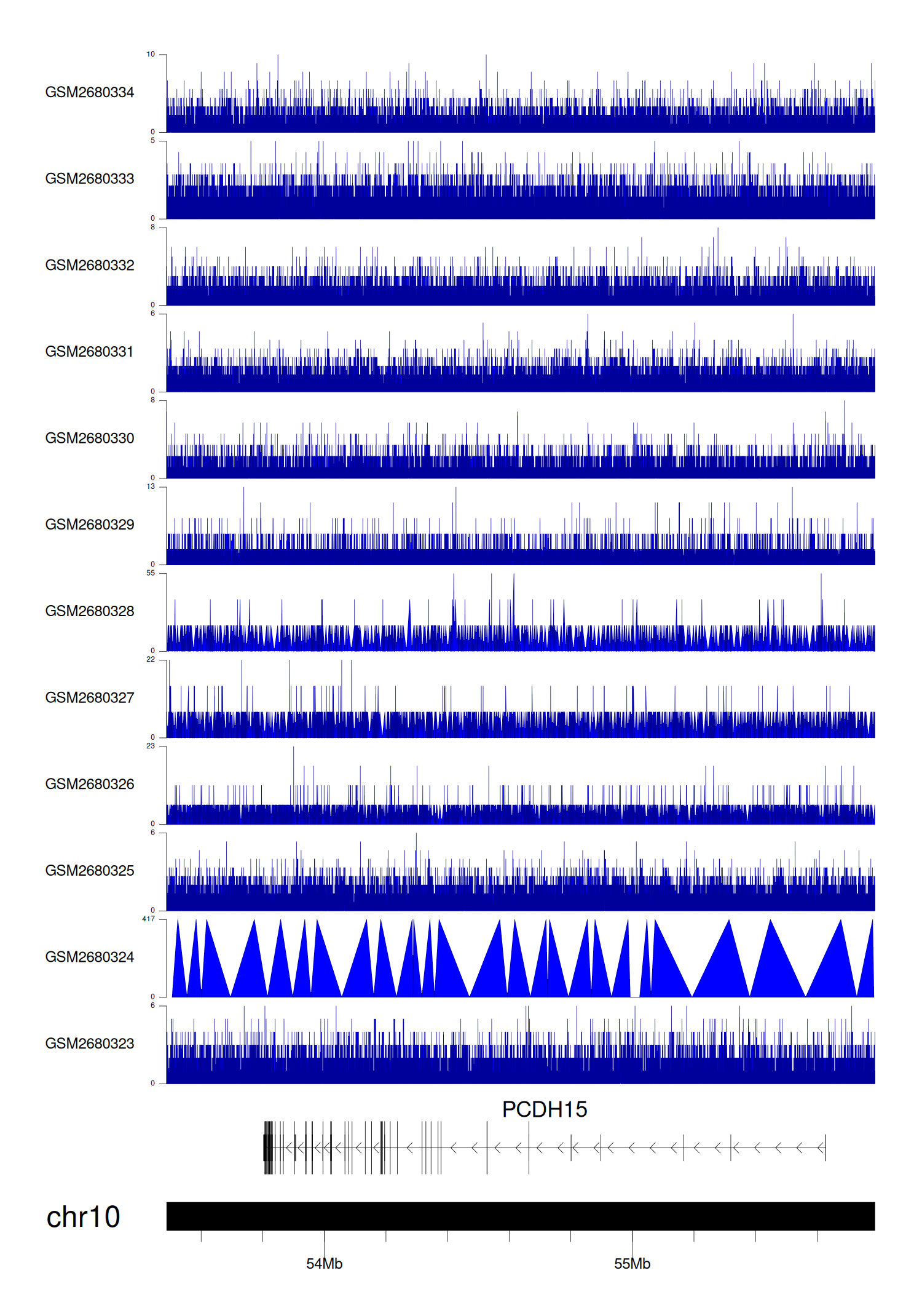
|
> Dataset: GSE131257 - PCDH15 peak across samples
|
Peak Plot
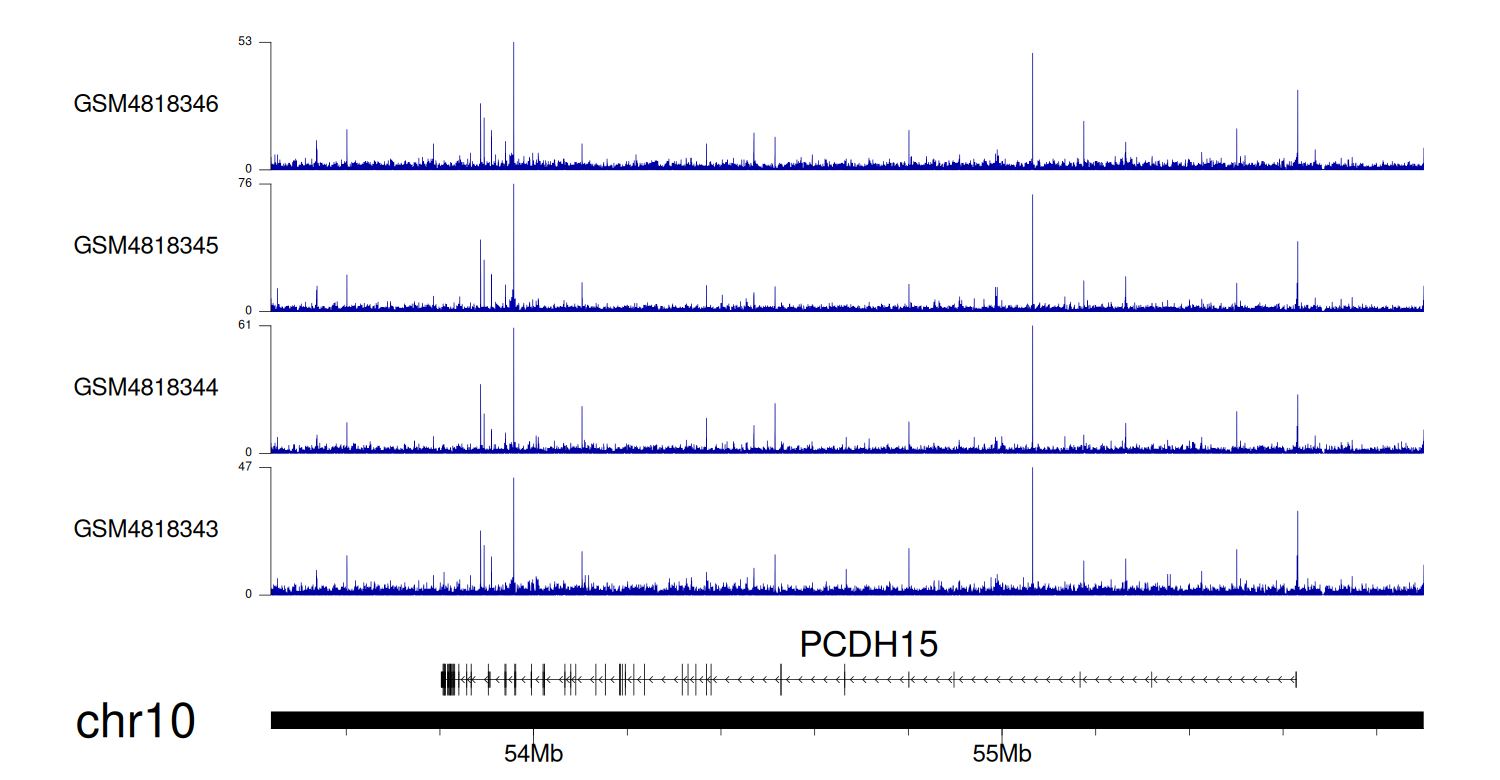
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information