Gene Information
|
Gene Name
|
PLG |
|
Gene ID
|
5340
|
|
Gene Full Name
|
plasminogen |
|
Gene Alias
|
HAE4 |
|
Transcripts
|
ENSG00000122194
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles;Secreted to blood; 
|
|
Membrane Info
|
Cancer-related genes, Candidate cardiovascular disease genes, Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Metabolic proteins, Plasma proteins, Predicted intracellular proteins, Predicted secreted proteins |
|
Uniport_ID
|
P00747
|
|
HGNC ID
|
HGNC:9071
|
|
OMIM ID
|
173350 |
|
Summary
|
The plasminogen protein encoded by this gene is a serine protease that circulates in blood plasma as an inactive zymogen and is converted to the active protease, plasmin, by several plasminogen activators such as tissue plasminogen activator (tPA), urokinase plasminogen activator (uPA), kallikrein, and factor XII (Hageman factor). The conversion of plasminogen to plasmin involves the cleavage of the peptide bond between Arg-561 and Val-562. Plasmin cleavage also releases the angiostatin protein which inhibits angiogenesis. Plasmin degrades many blood plasma proteins, including fibrin-containing blood clots. As a serine protease, plasmin cleaves many products in addition to fibrin such as fibronectin, thrombospondin, laminin, and von Willebrand factor. Plasmin is inactivated by proteins such as alpha-2-macroglobulin and alpha-2-antiplasmin in addition to inhibitors of the various plasminogen activators. Plasminogen also interacts with plasminogen receptors which results in the retention of plasmin on cell surfaces and in plasmin-induced cell signaling. The localization of plasminogen on cell surfaces plays a role in the degradation of extracellular matrices, cell migration, inflamation, wound healing, oncogenesis, metastasis, myogenesis, muscle regeneration, neurite outgrowth, and fibrinolysis. This protein may also play a role in acute respiratory distress syndrome (ARDS) which, in part, is caused by enhanced clot formation and the suppression of fibrinolysis. Compared to other mammals, the cluster of plasminogen-like genes to which this gene belongs has been rearranged in catarrhine primates. [provided by RefSeq, May 2020] |
Target gene [PLG] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1008660 |
chr6 |
20 |
0 |
0 |
20 |
View |
| 1009908 |
chr6 |
1 |
3 |
10 |
14 |
View |
| 1014246 |
chr6 |
4 |
0 |
0 |
4 |
View |
| 1043201 |
chr6 |
15 |
0 |
1 |
16 |
View |
Target gene [PLG] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - PLG expression across samples
|
Volcano Plot
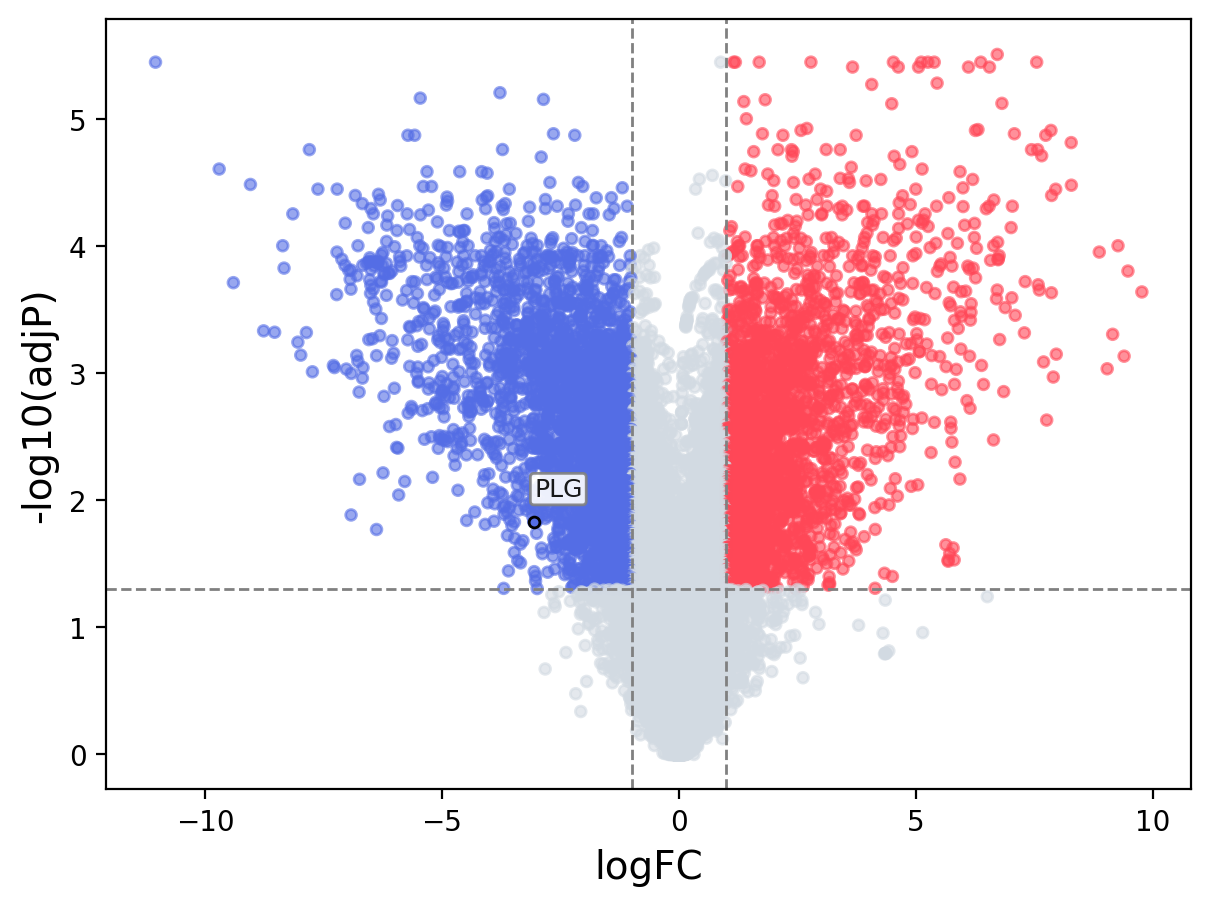
|
Bar Plot
|
> Dataset: GSE94660 - PLG expression across samples
|
Volcano Plot
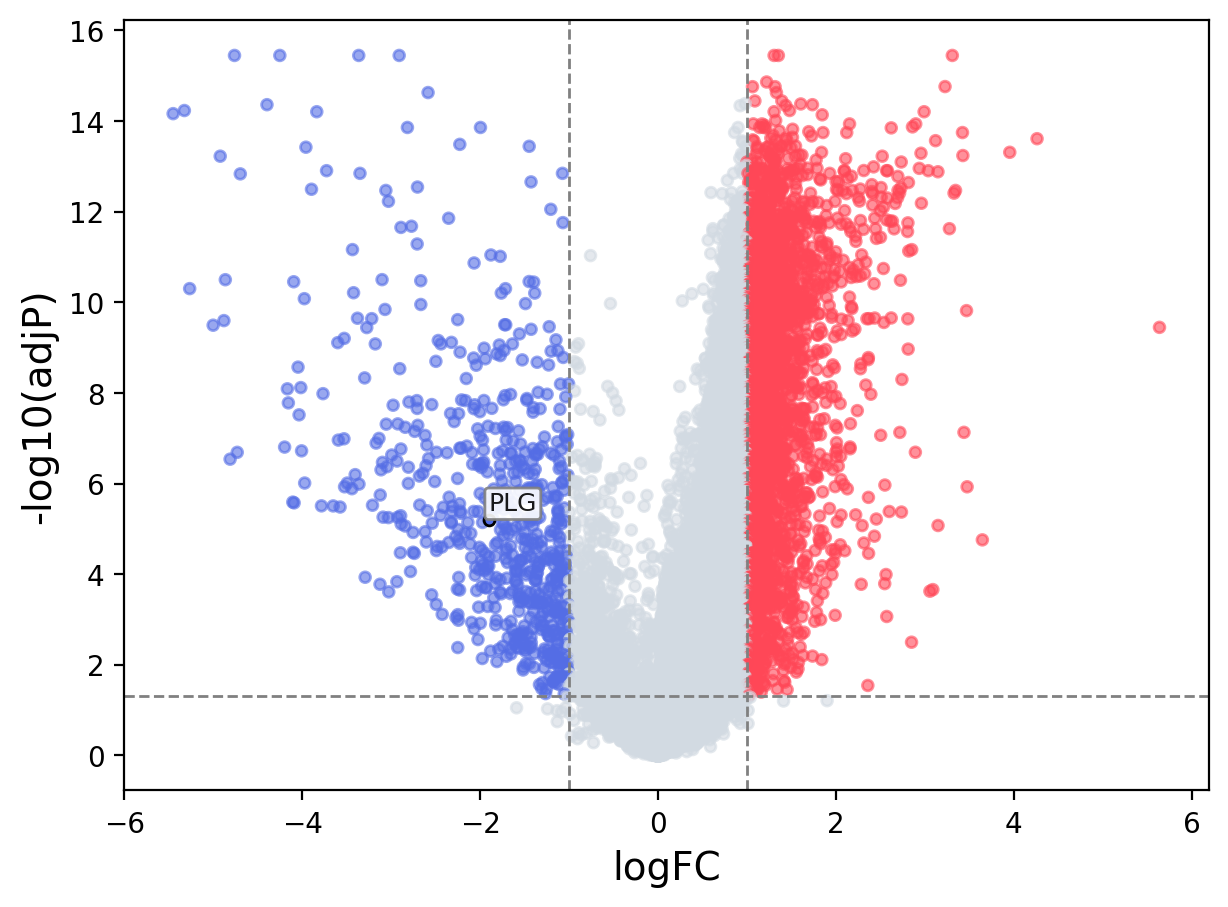
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information