Gene Information
|
Gene Name
|
PML |
|
Gene ID
|
5371
|
|
Gene Full Name
|
PML nuclear body scaffold |
|
Gene Alias
|
MYL|PP8675|RNF71|TRIM19 |
|
Transcripts
|
ENSG00000140464
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nuclear bodies; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins |
|
Uniport_ID
|
P29590
|
|
HGNC ID
|
HGNC:9113
|
|
OMIM ID
|
102578 |
|
Summary
|
The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. This phosphoprotein localizes to nuclear bodies where it functions as a transcription factor and tumor suppressor. Its expression is cell-cycle related and it regulates the p53 response to oncogenic signals. The gene is often involved in the translocation with the retinoic acid receptor alpha gene associated with acute promyelocytic leukemia (APL). Extensive alternative splicing of this gene results in several variations of the protein's central and C-terminal regions; all variants encode the same N-terminus. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008] |
Target gene [PML] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1039442 |
chr15 |
1 |
6 |
3 |
10 |
View |
| 1039443 |
chr15 |
1 |
7 |
3 |
11 |
View |
Target gene [PML] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
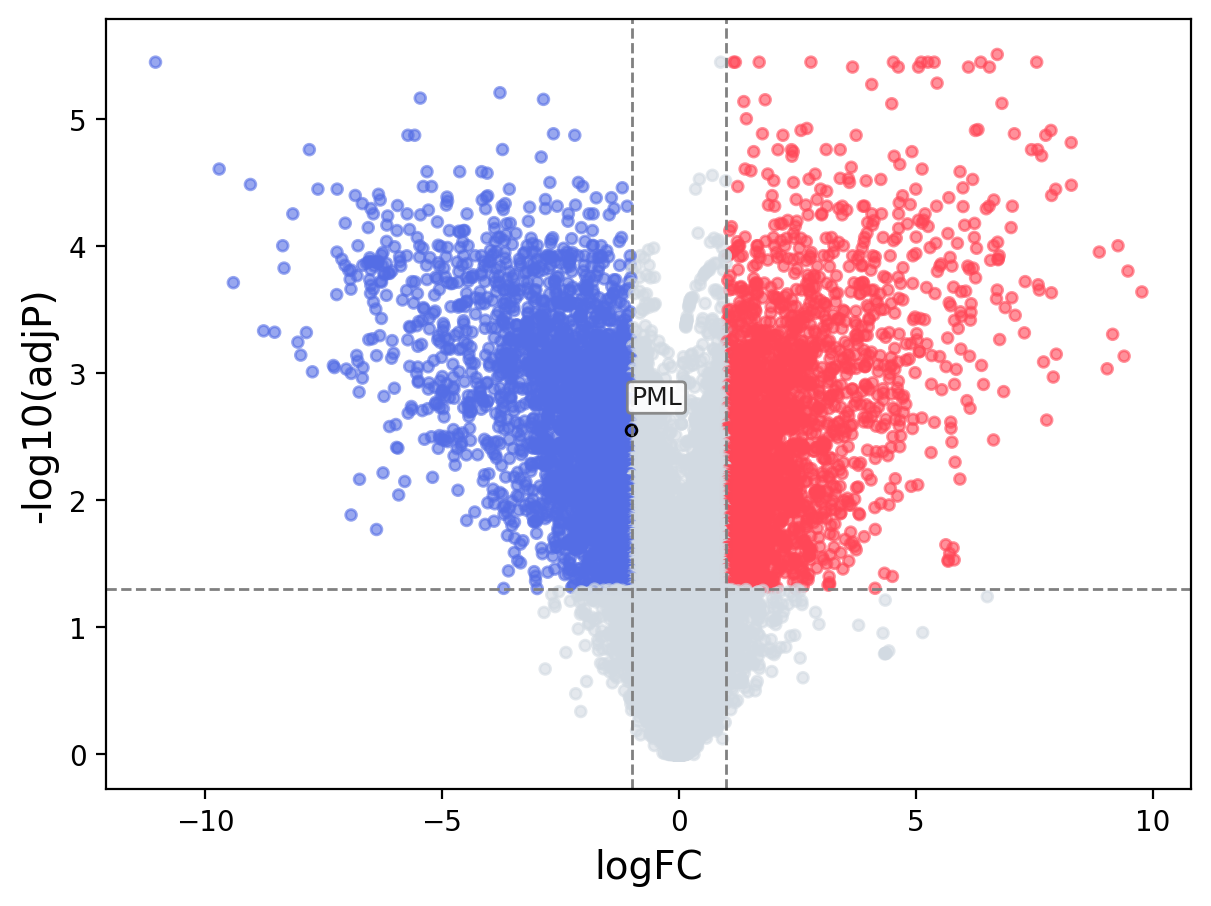
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - PML expression across samples
|
Volcano Plot
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information