Gene Information
|
Gene Name
|
PRDM16 |
|
Gene ID
|
63976
|
|
Gene Full Name
|
PR/SET domain 16 |
|
Gene Alias
|
CMD1LL|KMT8F|LVNC8|MEL1|PFM13 |
|
Transcripts
|
ENSG00000142611
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, Human disease related genes, Potential drug targets, Predicted intracellular proteins, Transcription factors |
|
Uniport_ID
|
Q9HAZ2
|
|
HGNC ID
|
HGNC:14000
|
|
OMIM ID
|
605557 |
|
Summary
|
The reciprocal translocation t(1;3)(p36;q21) occurs in a subset of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). This gene is located near the 1p36.3 breakpoint and has been shown to be specifically expressed in the t(1:3)(p36,q21)-positive MDS/AML. The protein encoded by this gene is a zinc finger transcription factor and contains an N-terminal PR domain. The translocation results in the overexpression of a truncated version of this protein that lacks the PR domain, which may play an important role in the pathogenesis of MDS and AML. Alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Jul 2008] |
Target gene [PRDM16] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1010375 |
chr1 |
11 |
1 |
0 |
12 |
View |
| 1015895 |
chr1 |
2 |
0 |
0 |
2 |
View |
| 1024718 |
chr1 |
5 |
0 |
2 |
7 |
View |
| 1024766 |
chr1 |
3 |
5 |
2 |
10 |
View |
Target gene [PRDM16] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - PRDM16 peak across samples
|
Peak Plot
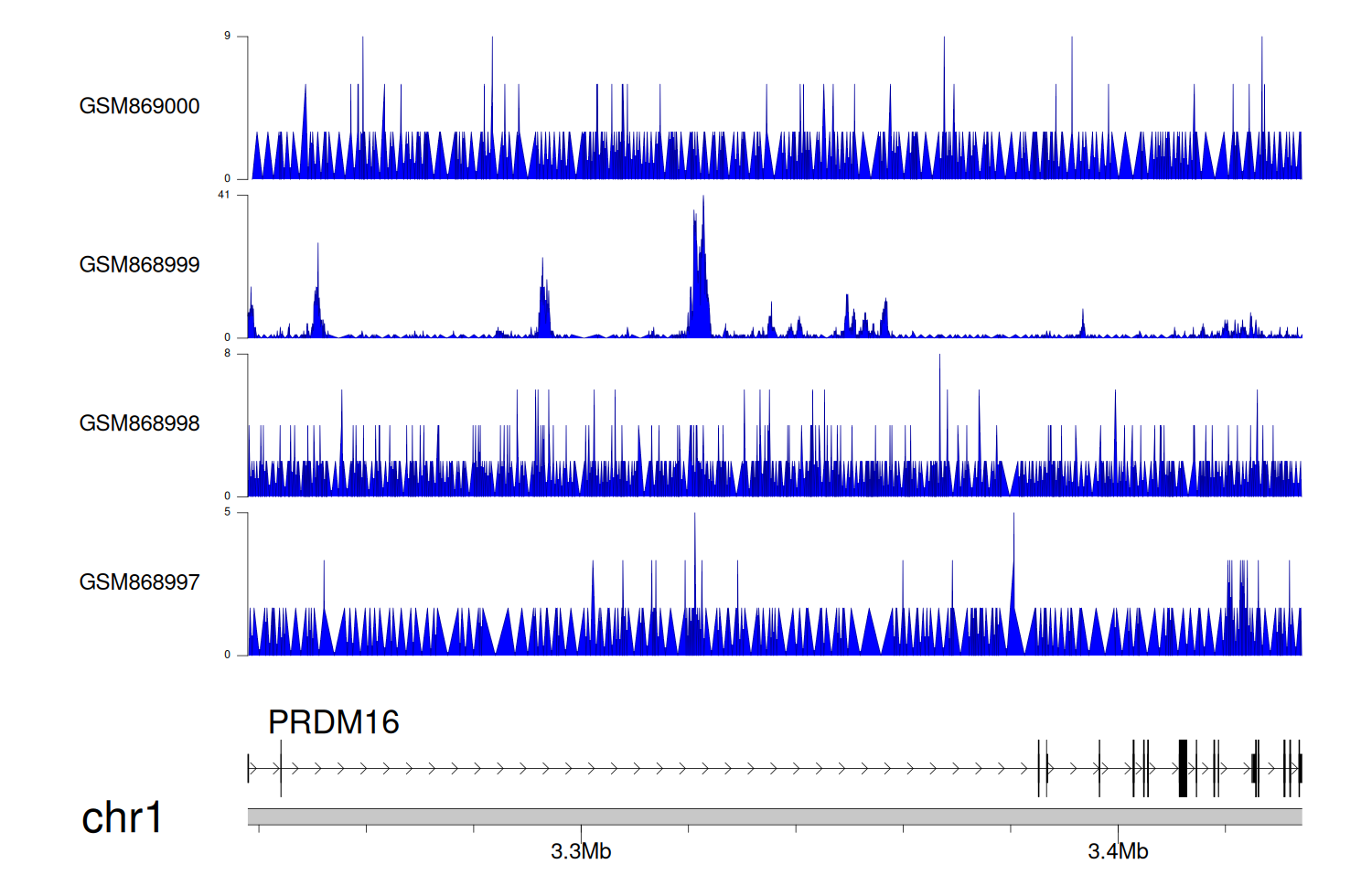
|
> Dataset: GSE68402 - PRDM16 peak across samples
|
Peak Plot
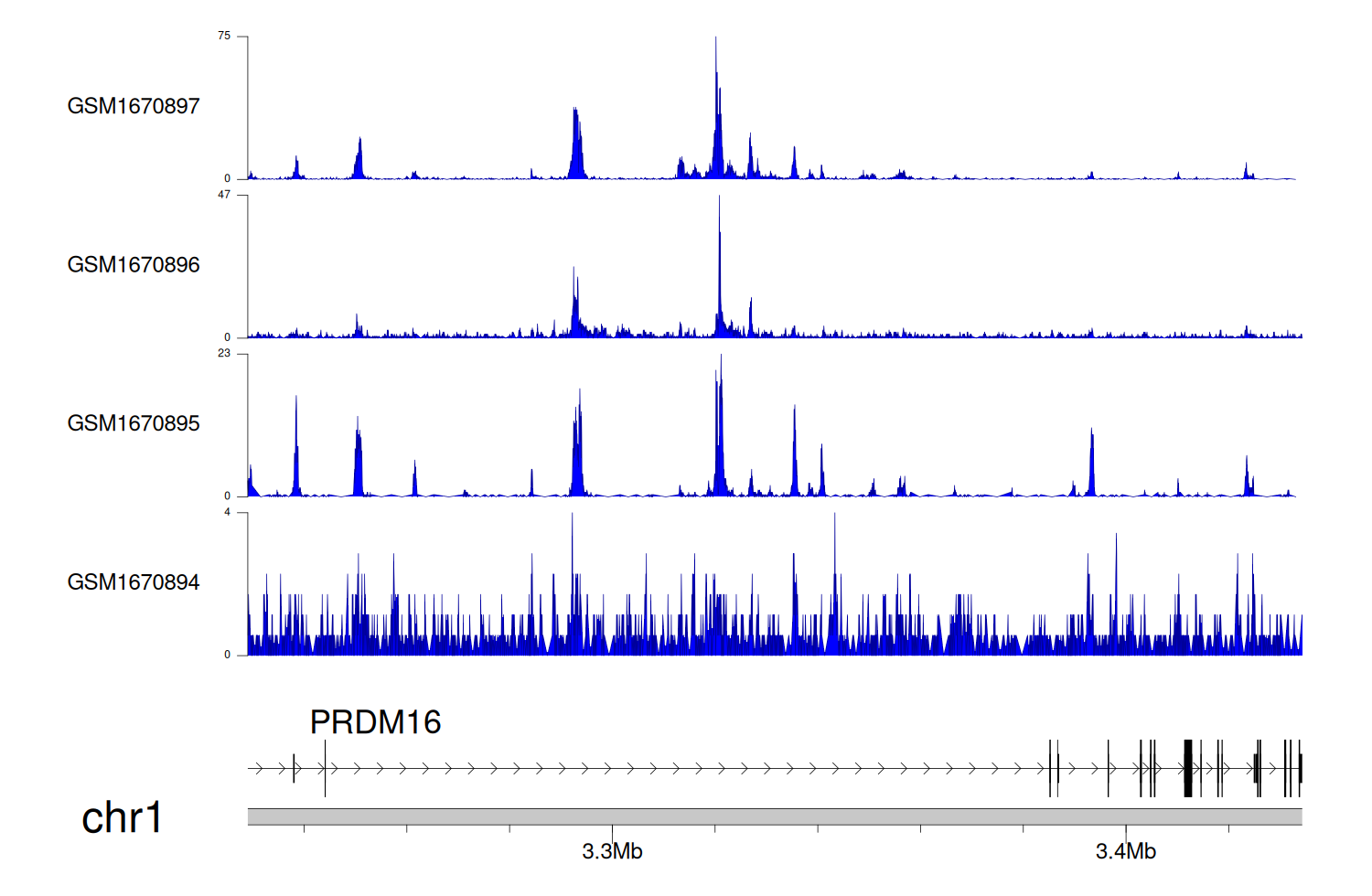
|
> Dataset: GSE270130 - PRDM16 peak across samples
|
Peak Plot
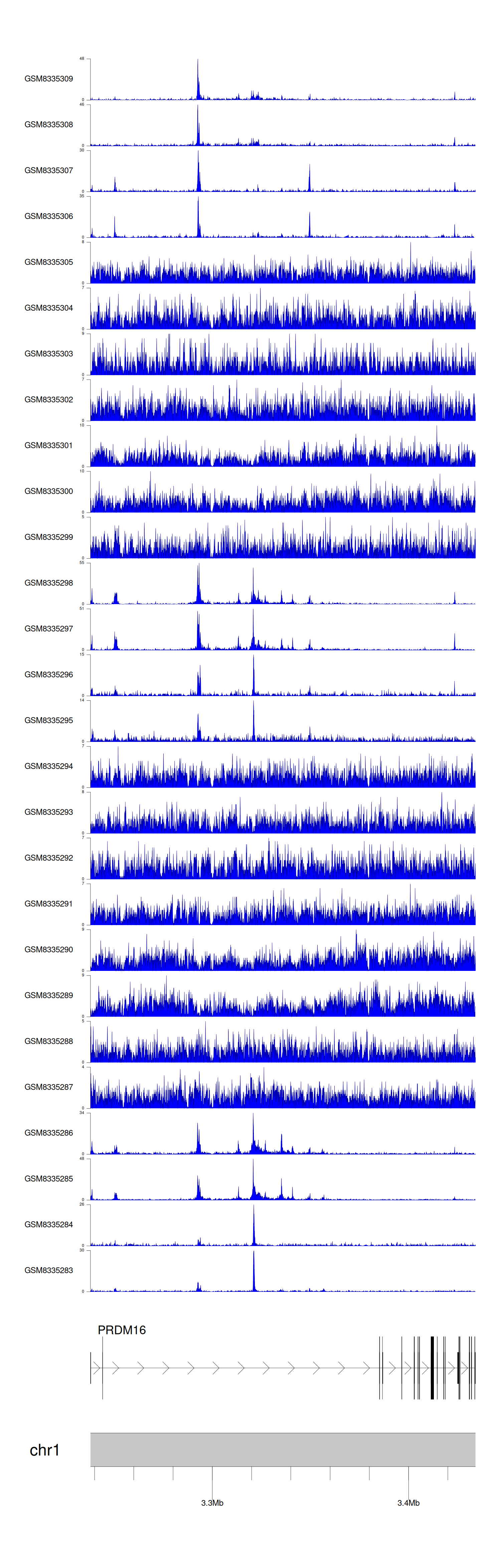
|
> Dataset: GSE131257 - PRDM16 peak across samples
|
Peak Plot
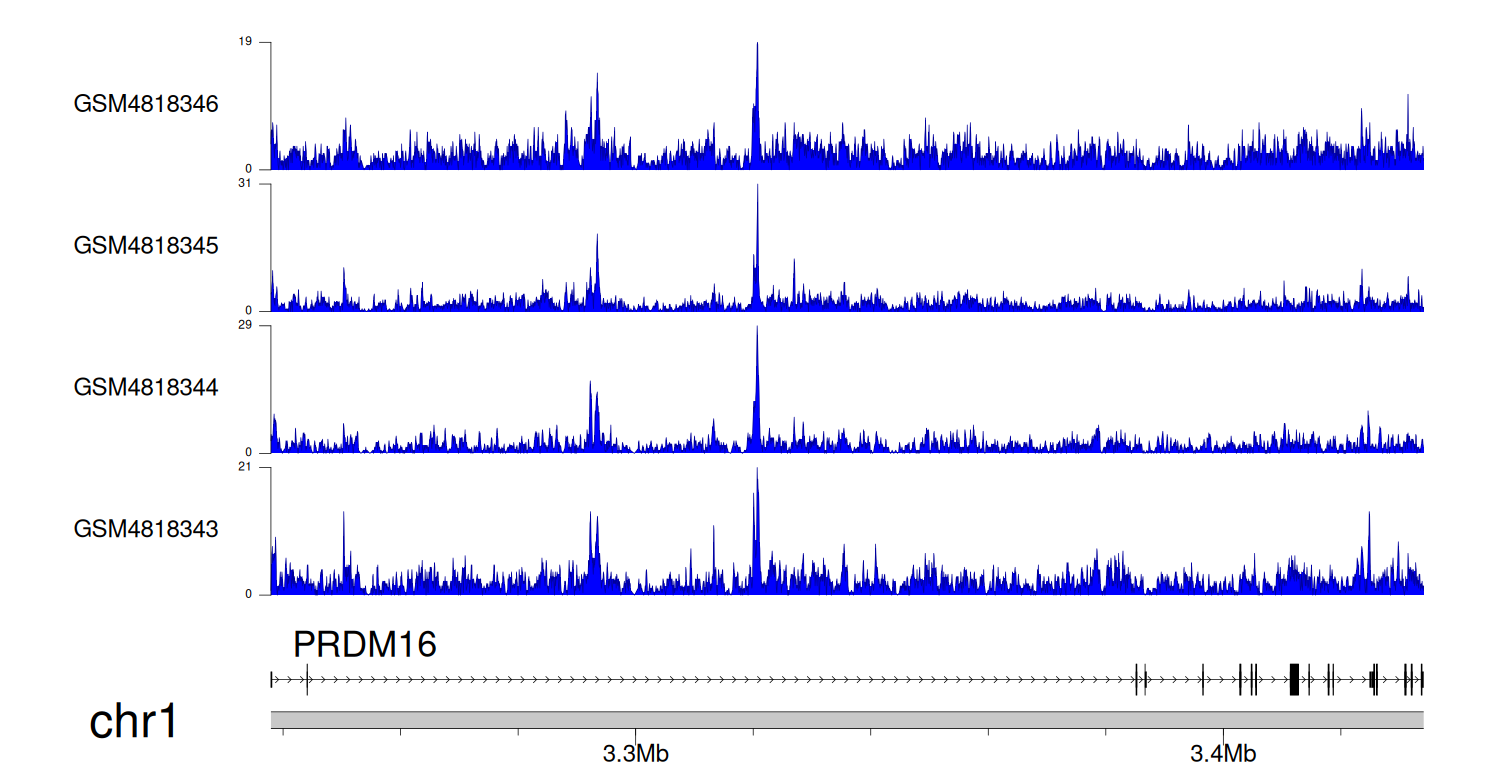
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information