Gene Information
|
Gene Name
|
PREX2 |
|
Gene ID
|
80243
|
|
Gene Full Name
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2 |
|
Gene Alias
|
DEP.2|DEPDC2|P-REX2|PPP1R129 |
|
Transcripts
|
ENSG00000046889
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Endoplasmic reticulum; 
|
|
Membrane Info
|
Cancer-related genes, Plasma proteins, Predicted intracellular proteins |
|
Uniport_ID
|
Q70Z35
|
|
HGNC ID
|
HGNC:22950
|
|
OMIM ID
|
612139 |
|
Summary
|
The protein encoded by this gene belongs to the phosphatidylinositol 3,4,5-trisphosphate (PIP3)-dependent Rac exchanger (PREX) family, which are Dbl-type guanine-nucleotide exchange factors for Rac family small G proteins. Structural domains of this protein include the catalytic diffuse B-cell lymphoma homology and pleckstrin homology (DHPH) domain, two disheveled, EGL-10, and pleckstrin homology (DEP) domains, two PDZ domains, and a C-terminal inositol polyphosphate-4 phosphatase (IP4P) domain that is found in one of the isoforms. This protein facilitates the exchange of GDP for GTP on Rac1, allowing the GTP-bound Rac1 to activate downstream effectors. Studies also show that the pleckstrin homology domain of this protein interacts with the phosphatase and tensin homolog (PTEN) gene product to inhibit PTEN phosphatase activity, thus activating the phosphoinositide-3 kinase (PI3K) signaling pathway. Conversely, the PTEN gene product has also been shown to inhibit the GEF activity of this protein. This gene plays a role in insulin-signaling pathways, and either mutations or overexpression of this gene have been observed in some cancers. [provided by RefSeq, Apr 2016] |
Target gene [PREX2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
Target gene [PREX2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE68402 - PREX2 peak across samples
|
Peak Plot
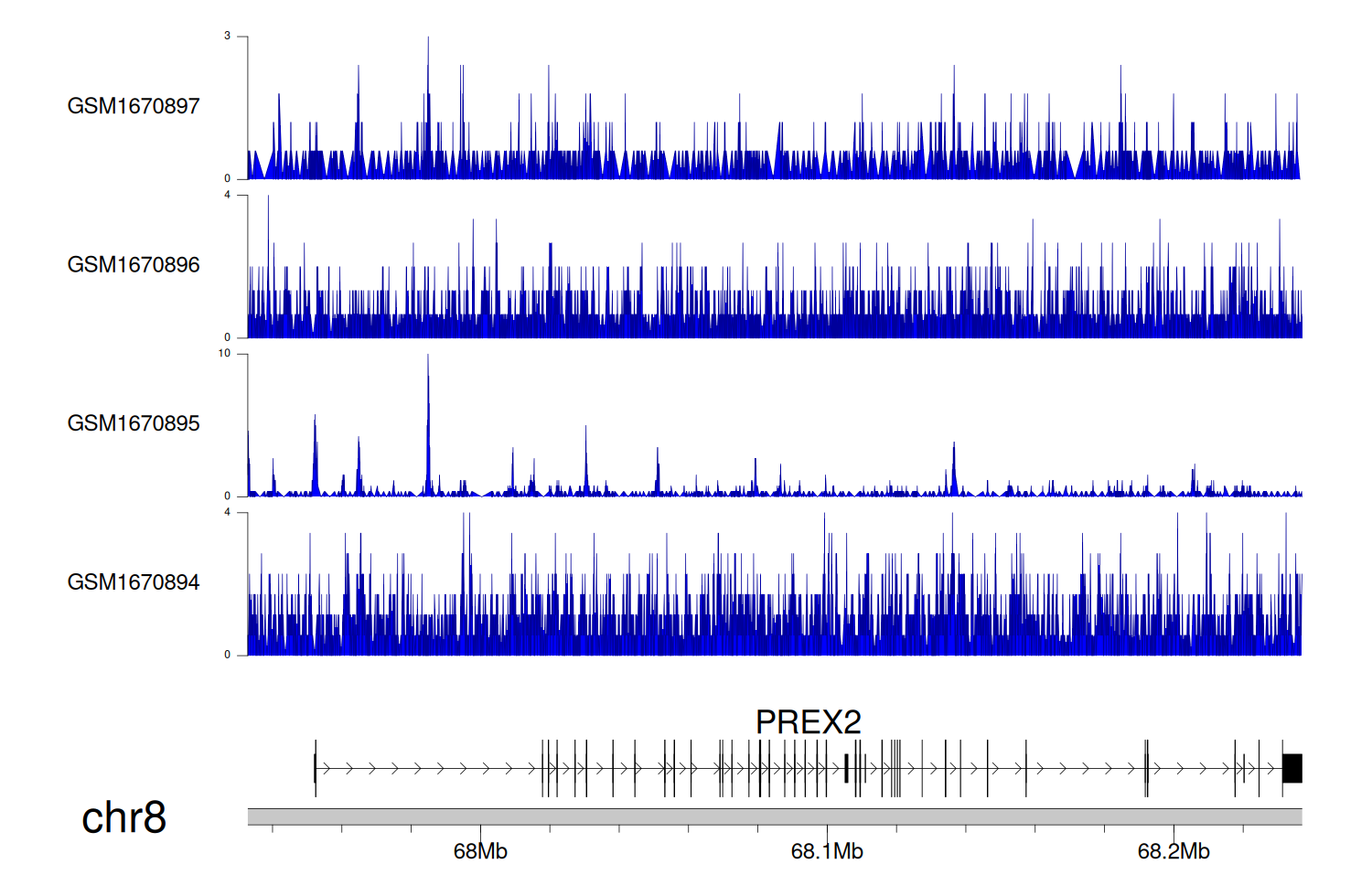
|
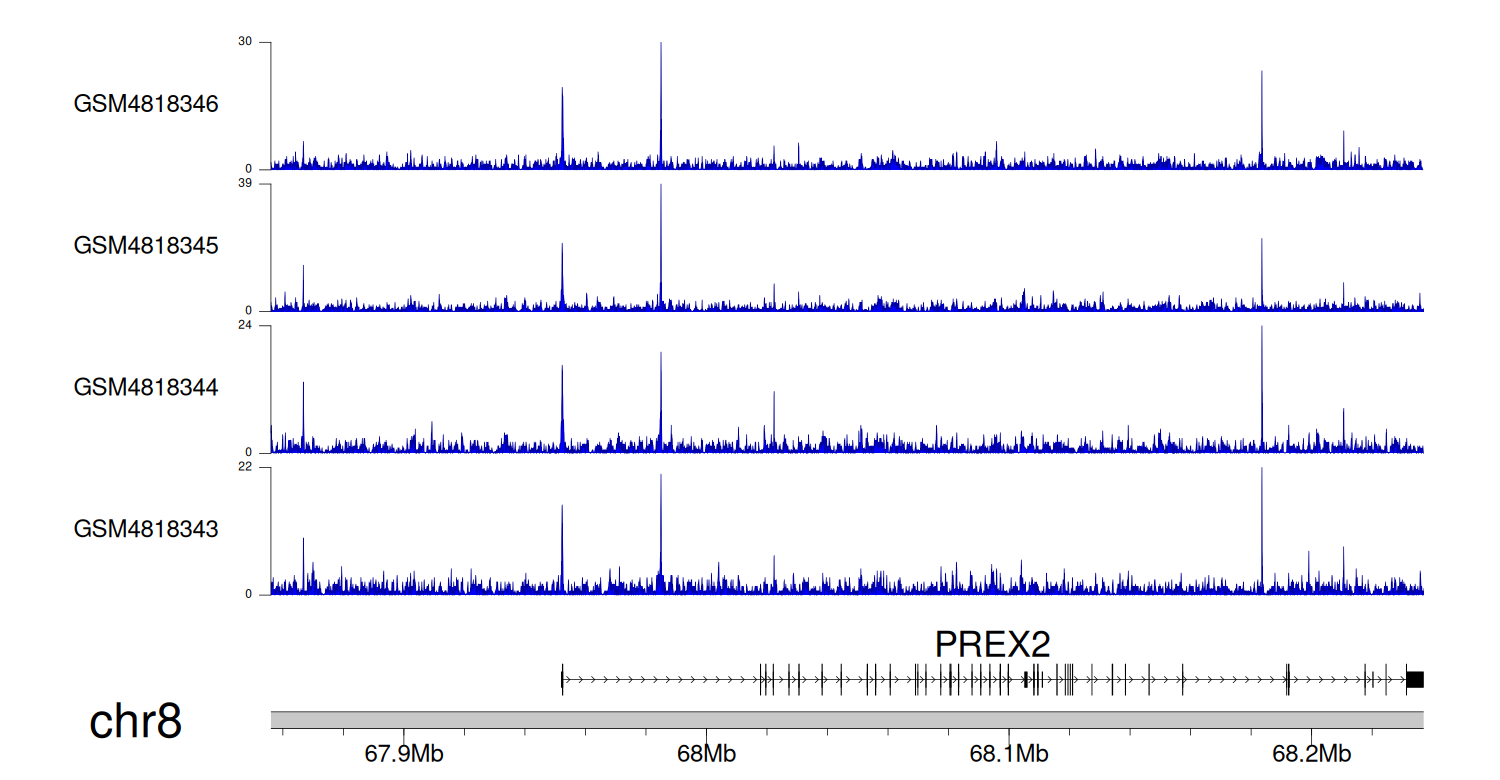
> Dataset: GSE270130 - PREX2 peak across samples
|
Peak Plot
|
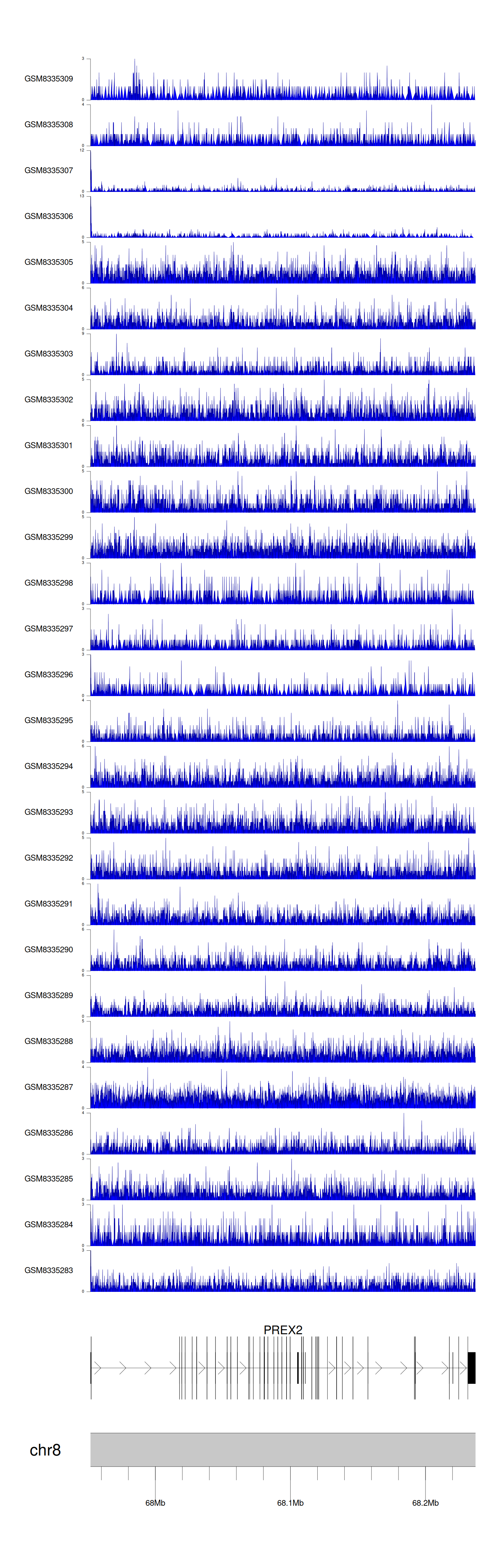
> Dataset: GSE131257 - PREX2 peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information