Gene Information
|
Gene Name
|
PRPH2 |
|
Gene ID
|
5961
|
|
Gene Full Name
|
peripherin 2 |
|
Gene Alias
|
AOFMD|AVMD|CACD2|DS|MDBS1|PRPH|RDS|RP7|TSPAN22|rd2 |
|
Transcripts
|
ENSG00000112619
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Potential drug targets, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
P23942
|
|
HGNC ID
|
HGNC:9942
|
|
OMIM ID
|
179605 |
|
Summary
|
The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of four hydrophobic domains. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. This encoded protein is a cell surface glycoprotein found in the outer segment of both rod and cone photoreceptor cells. It may function as an adhesion molecule involved in stabilization and compaction of outer segment disks or in the maintenance of the curvature of the rim. This protein is essential for disk morphogenesis. Defects in this gene are associated with both central and peripheral retinal degenerations. Some of the various phenotypically different disorders are autosomal dominant retinitis pigmentosa, progressive macular degeneration, macular dystrophy and retinitis pigmentosa digenic. [provided by RefSeq, Jul 2008] |
Target gene [PRPH2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1015703 |
chr6 |
1 |
1 |
0 |
2 |
View |
Target gene [PRPH2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - PRPH2 peak across samples
|
Peak Plot
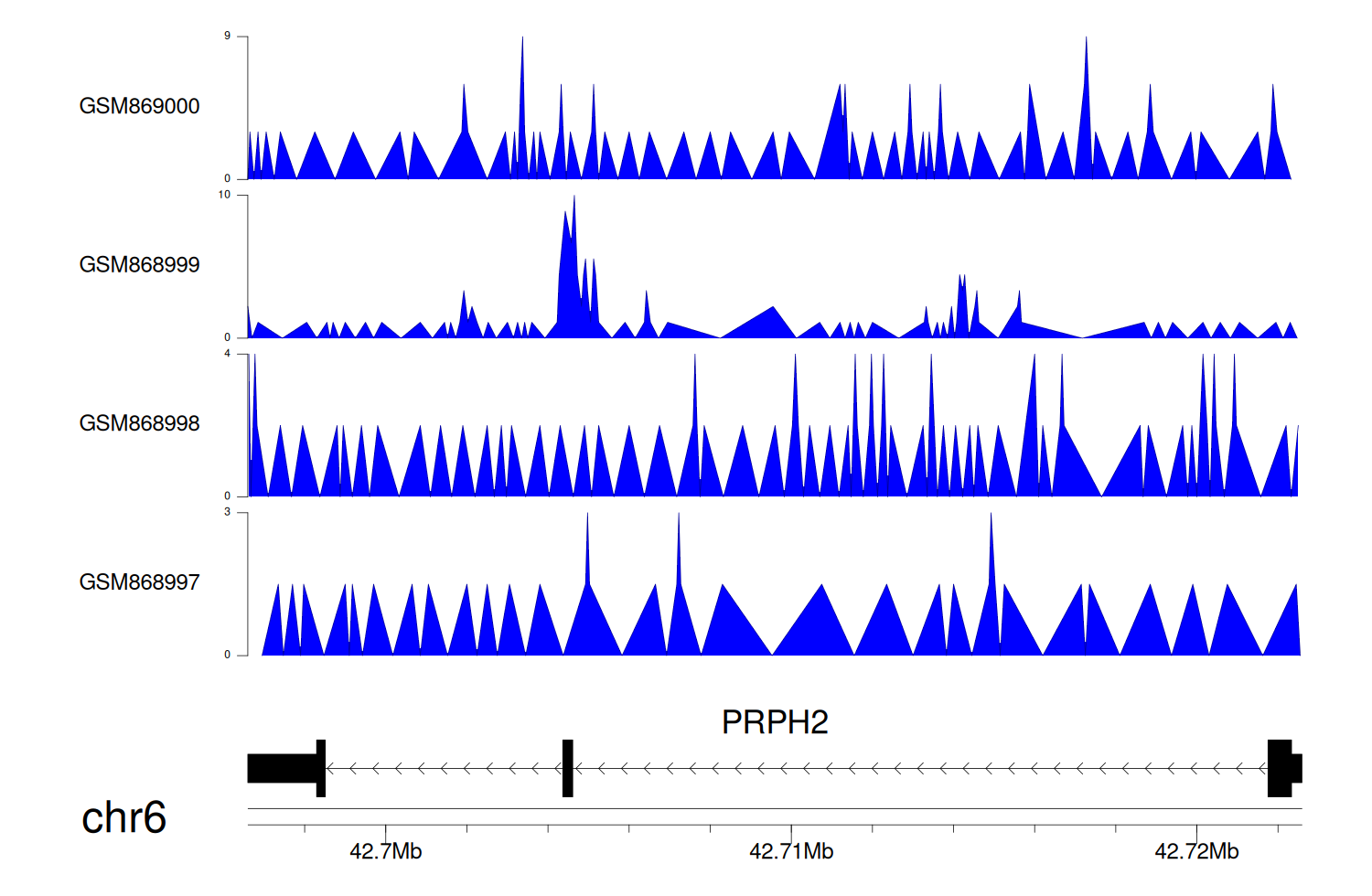
|
> Dataset: GSE68402 - PRPH2 peak across samples
|
Peak Plot
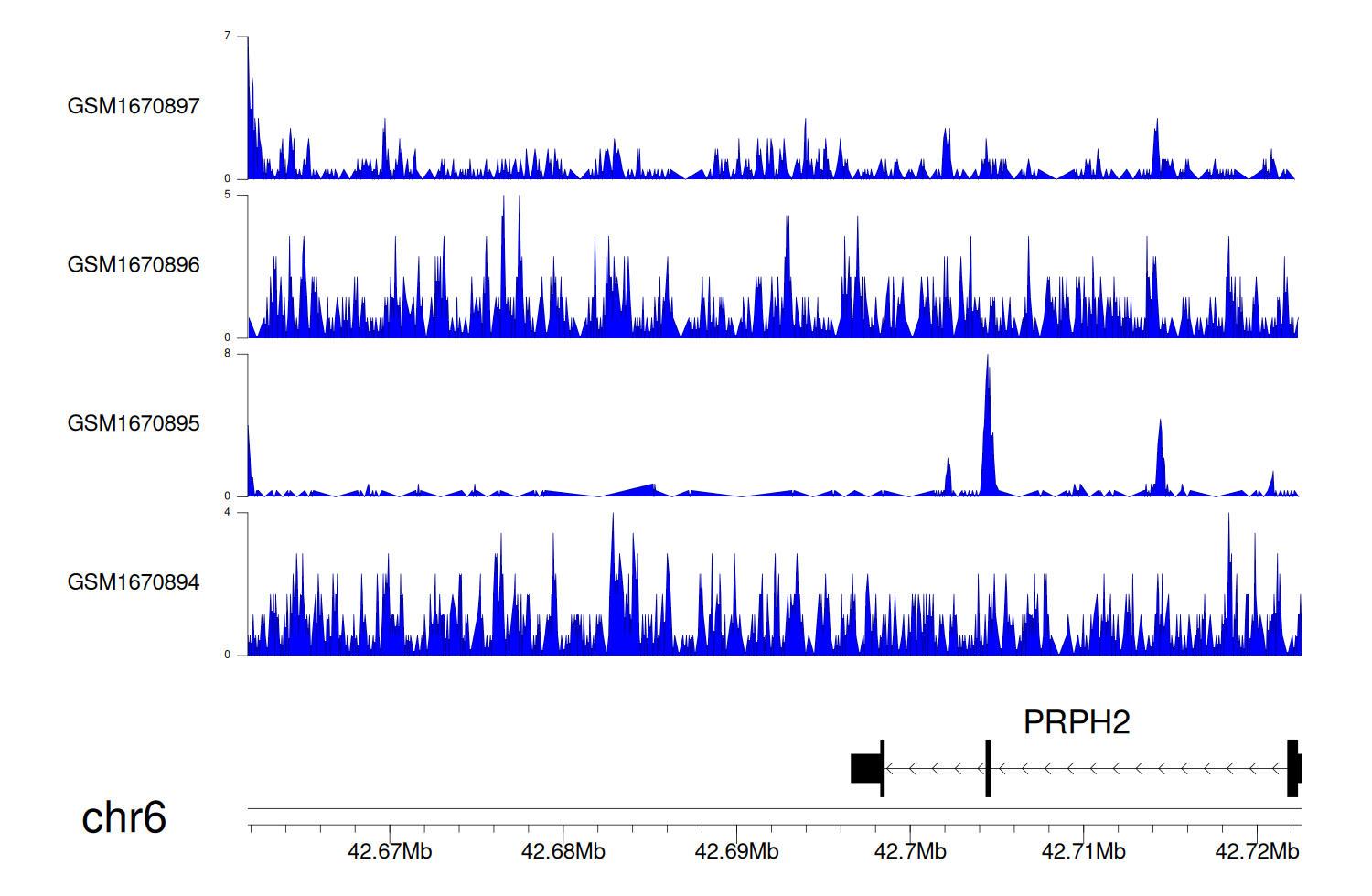
|
> Dataset: GSE270130 - PRPH2 peak across samples
|
Peak Plot
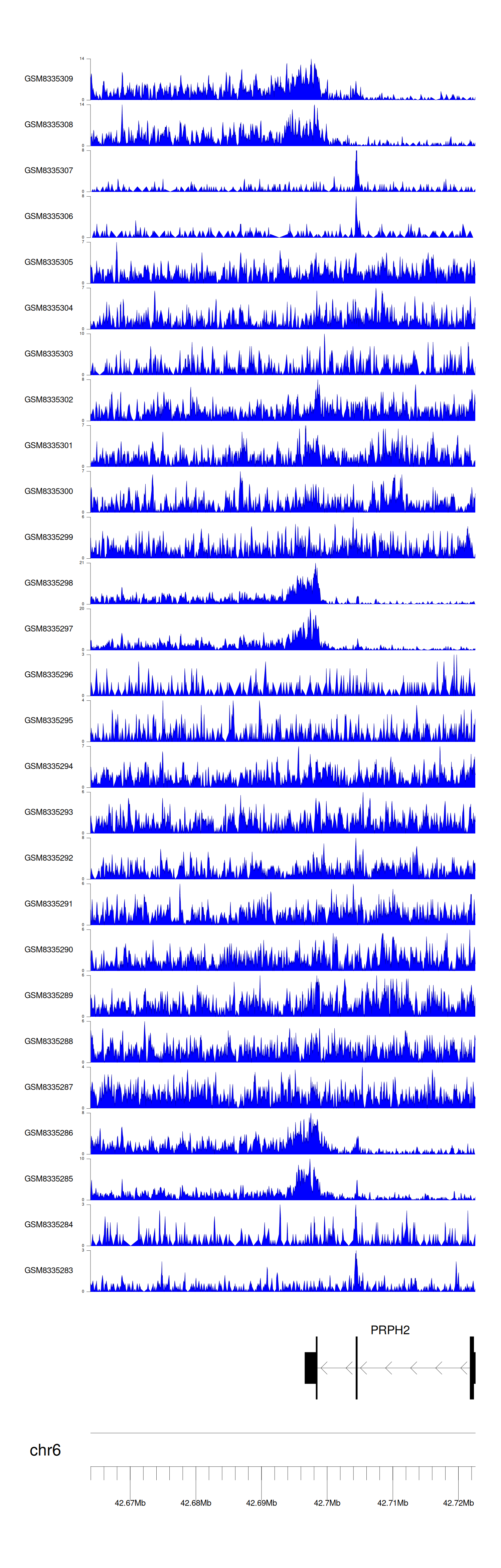
|
> Dataset: GSE131257 - PRPH2 peak across samples
|
Peak Plot
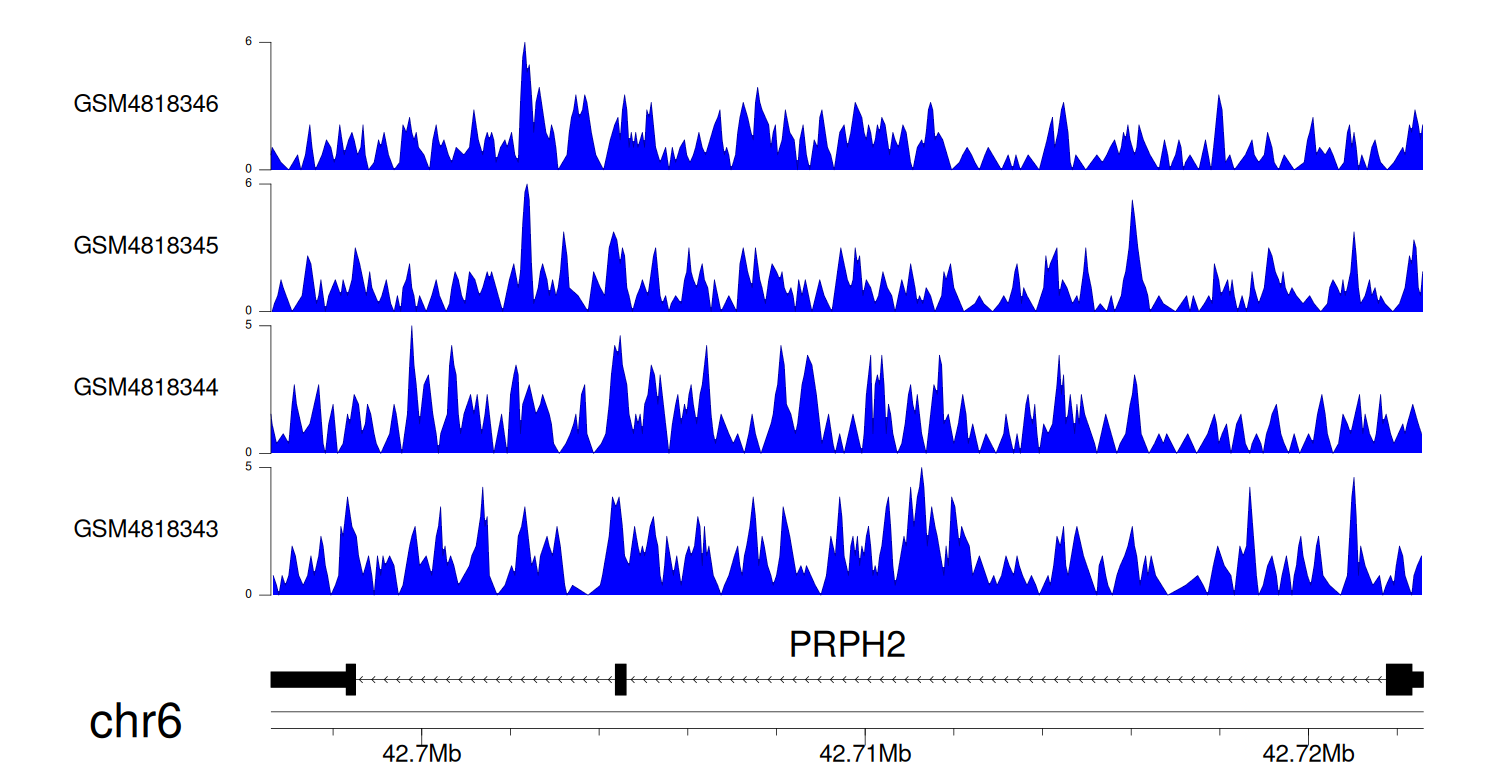
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information