Gene Information
|
Gene Name
|
SELENON |
|
Gene ID
|
57190
|
|
Gene Full Name
|
selenoprotein N |
|
Gene Alias
|
CFTD|CMYO3|CMYP3|MDRS1|RSMD1|RSS|SELN|SEPN1 |
|
Transcripts
|
ENSG00000162430
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Cytosol;Intracellular and membrane; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins |
|
Uniport_ID
|
Q9NZV5
|
|
HGNC ID
|
HGNC:15999
|
|
OMIM ID
|
606210 |
|
Summary
|
This gene encodes a glycoprotein that is localized in the endoplasmic reticulum. It plays an important role in cell protection against oxidative stress, and in the regulation of redox-related calcium homeostasis. Mutations in this gene are associated with early onset muscle disorders, referred to as SEPN1-related myopathy. SEPN1-related myopathy consists of 4 autosomal recessive disorders, originally thought to be separate entities: rigid spine muscular dystrophy (RSMD1), the classical form of multiminicore disease, desmin related myopathy with Mallory-body like inclusions, and congenital fiber-type disproportion (CFTD). This protein is a selenoprotein, containing the rare amino acid selenocysteine (Sec). Sec is encoded by the UGA codon, which normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, designated the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon, rather than as a stop signal. A second stop-codon redefinition element (SRE) adjacent to the UGA codon has been identified in this gene (PMID:15791204). SRE is a phylogenetically conserved stem-loop structure that stimulates readthrough at the UGA codon, and augments the Sec insertion efficiency by SECIS. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Dec 2016] |
Target gene [SELENON] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1041661 |
chr1 |
72 |
33 |
53 |
158 |
View |
Target gene [SELENON] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
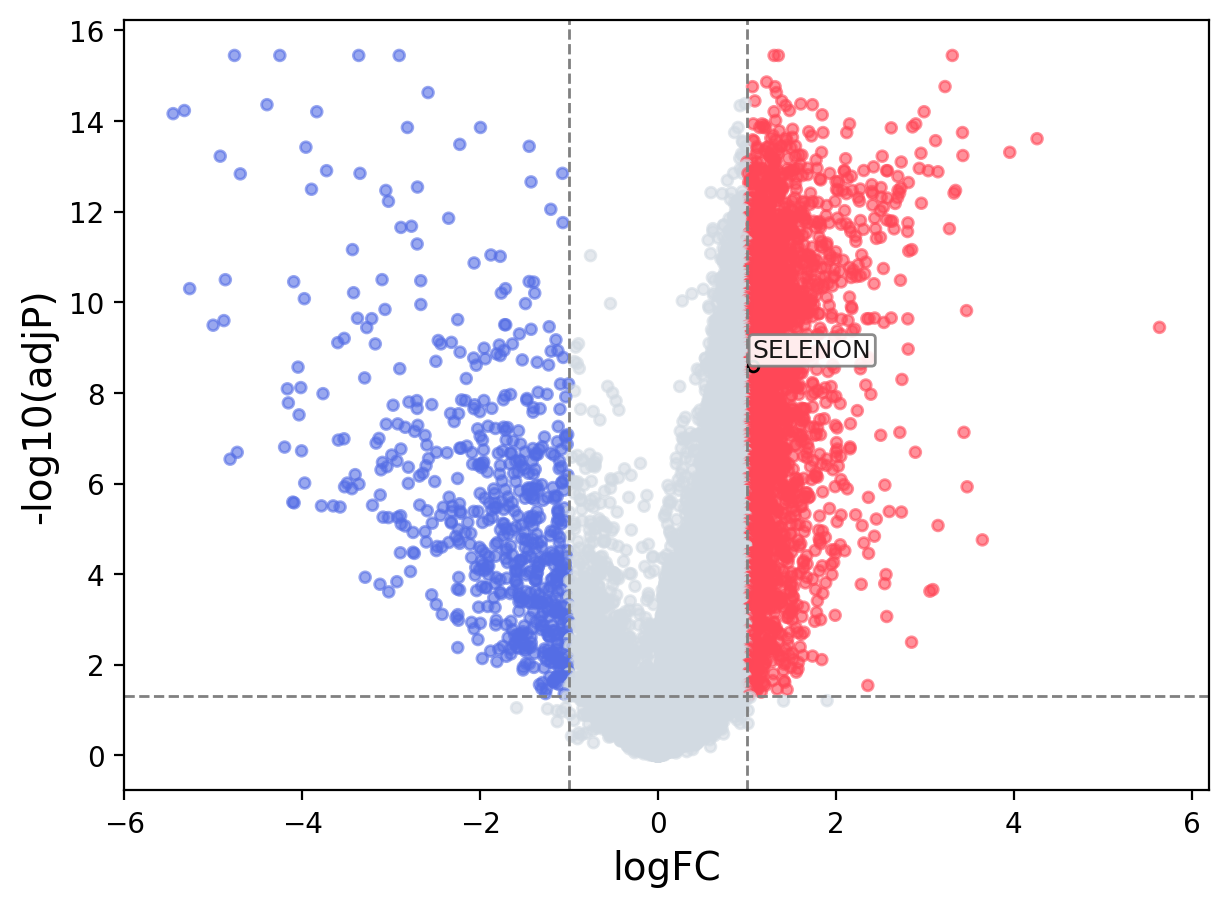
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE94660 - SELENON expression across samples
|
Volcano Plot
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information