Gene Information
|
Gene Name
|
SORCS3 |
|
Gene ID
|
22986
|
|
Gene Full Name
|
sortilin related VPS10 domain containing receptor 3 |
|
Gene Alias
|
SORCS |
|
Transcripts
|
ENSG00000156395
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
Q9UPU3
|
|
HGNC ID
|
HGNC:16699
|
|
OMIM ID
|
606285 |
|
Summary
|
This gene encodes a type-I receptor transmembrane protein that is a member of the vacuolar protein sorting 10 receptor family. Proteins of this family are defined by a vacuolar protein sorting 10 domain at the N-terminus. The N-terminal segment of this domain has a consensus motif for proprotein convertase processing, and the C-terminal segment of this domain is characterized by ten conserved cysteine residues. The vacuolar protein sorting 10 domain is followed by a leucine-rich segment, a transmembrane domain, and a short C-terminal cytoplasmic domain that interacts with adaptor molecules. The transcript is expressed at high levels in the brain, and candidate gene studies suggest that genetic variation in this gene is associated with Alzheimer's disease. Consistent with this observation, knockdown of the gene in cell culture results in an increase in amyloid precursor protein processing. [provided by RefSeq, Dec 2014] |
Target gene [SORCS3] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1012175 |
chr10 |
1 |
2 |
4 |
7 |
View |
| 1013083 |
chr10 |
5 |
0 |
0 |
5 |
View |
| 1015181 |
chr10 |
2 |
0 |
0 |
2 |
View |
| 1023081 |
chr10 |
5 |
0 |
0 |
5 |
View |
| 1041851 |
chr10 |
8 |
1 |
1 |
10 |
View |
| 1041852 |
chr10 |
200 |
7 |
0 |
207 |
View |
Target gene [SORCS3] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE68402 - SORCS3 peak across samples
|
Peak Plot
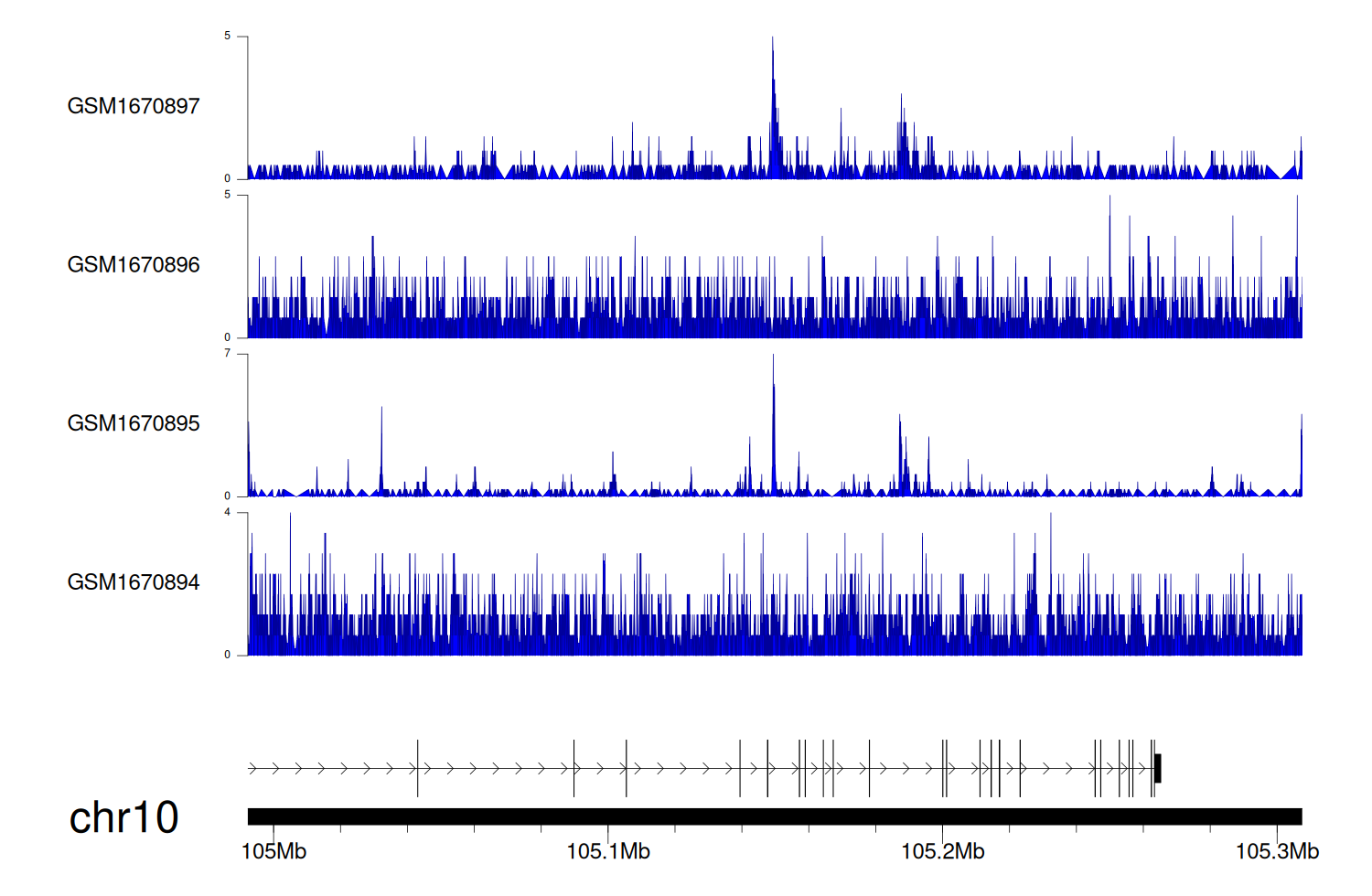
|
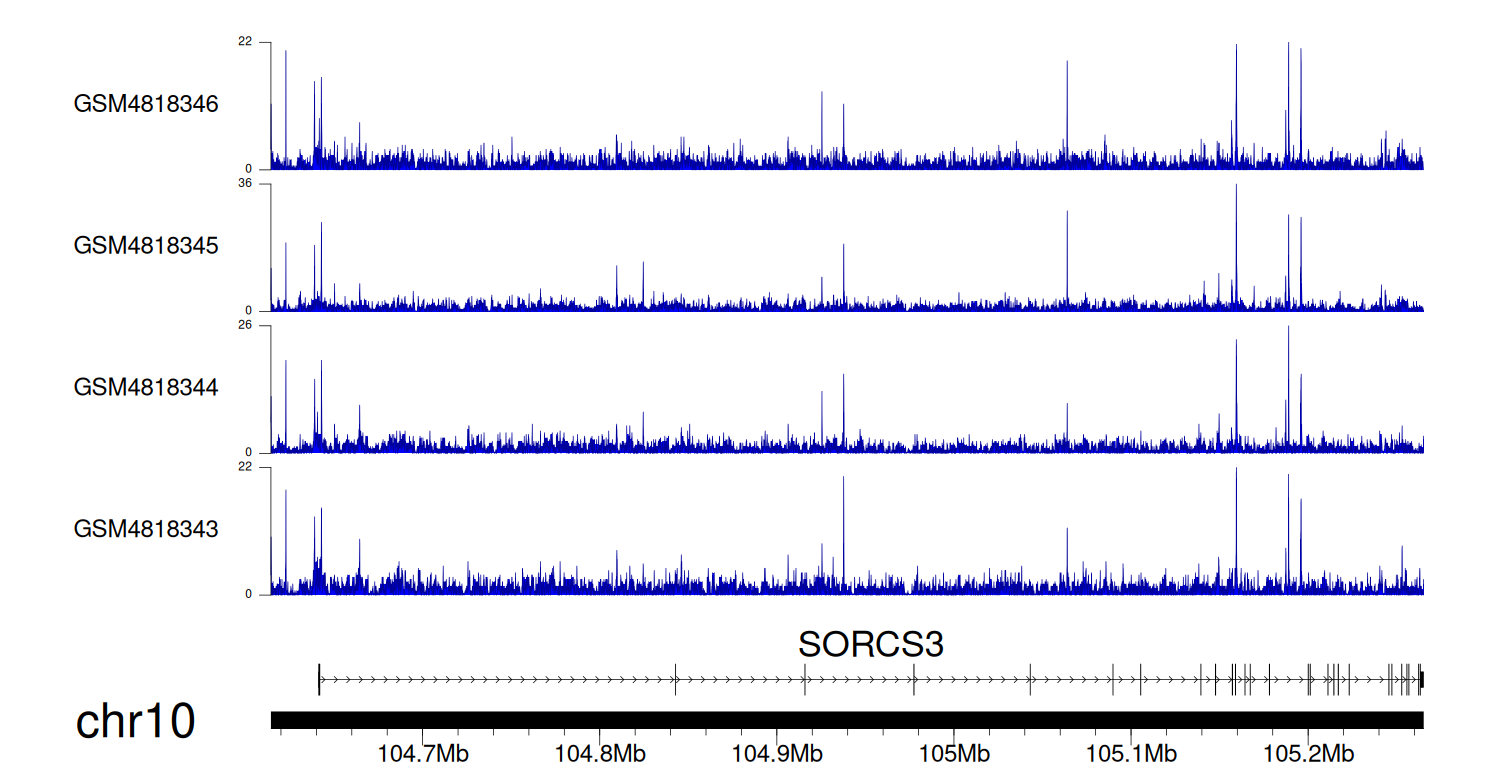
> Dataset: GSE270130 - SORCS3 peak across samples
|
Peak Plot
|
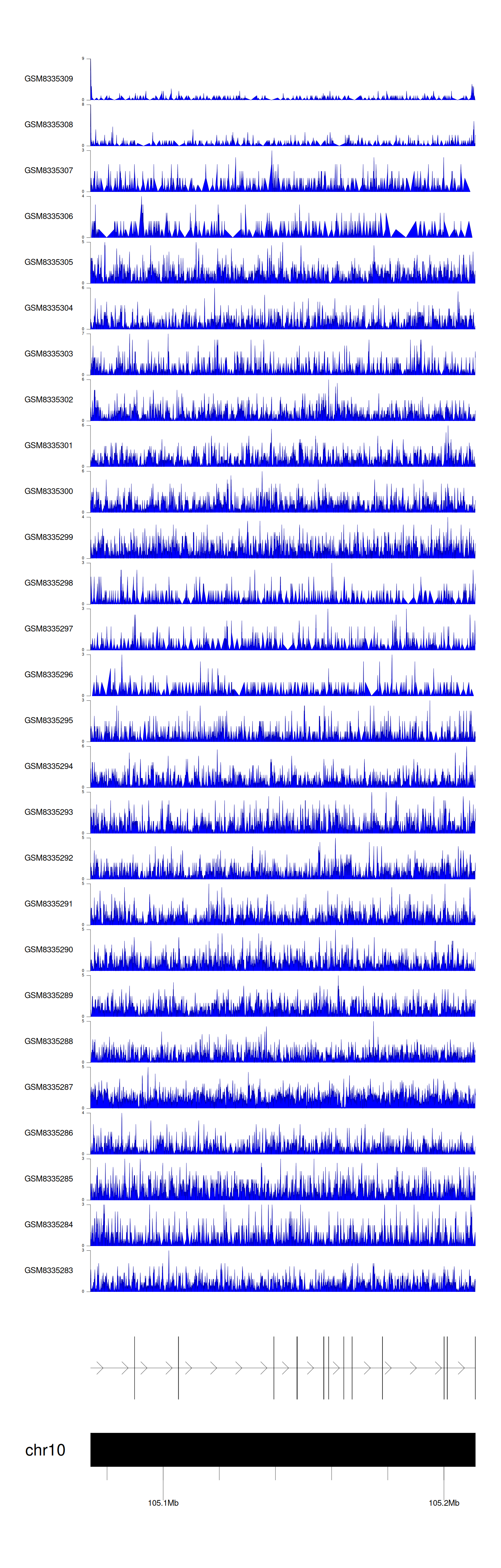
> Dataset: GSE100400 - SORCS3 peak across samples
|
Peak Plot
|
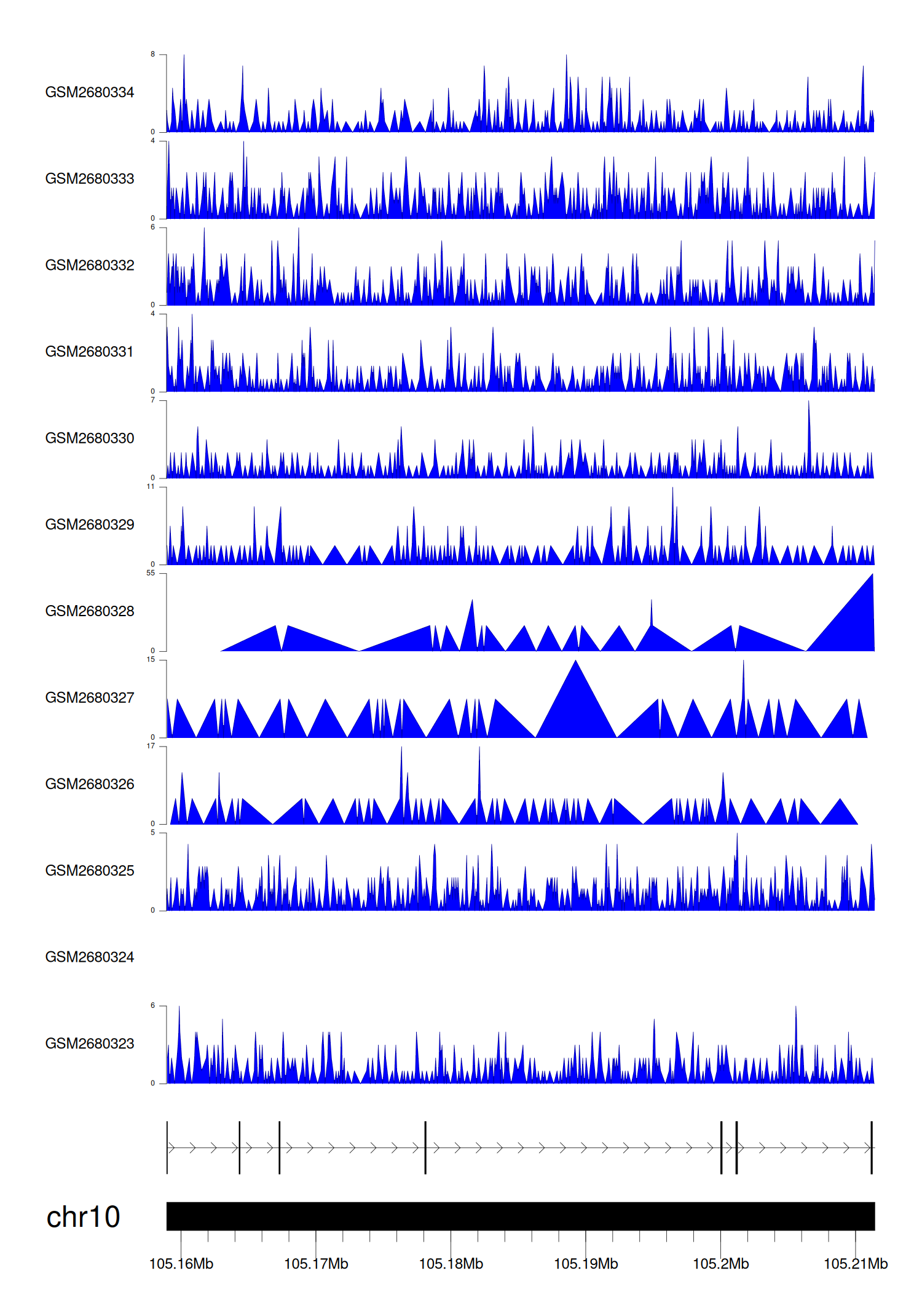
> Dataset: GSE131257 - SORCS3 peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information