Gene Information
|
Gene Name
|
TTN |
|
Gene ID
|
7273
|
|
Gene Full Name
|
titin |
|
Gene Alias
|
CMD1G|CMH9|CMPD4|CMYO5|CMYP5|EOMFC|HMERF|LGMD2J|LGMDR10|MYLK5|SALMY|TMD |
|
Transcripts
|
ENSG00000155657
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Plasma proteins, Potential drug targets, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
Q8WZ42
|
|
HGNC ID
|
HGNC:12403
|
|
OMIM ID
|
188840 |
|
Summary
|
This gene encodes a large abundant protein of striated muscle. The product of this gene is divided into two regions, a N-terminal I-band and a C-terminal A-band. The I-band, which is the elastic part of the molecule, contains two regions of tandem immunoglobulin domains on either side of a PEVK region that is rich in proline, glutamate, valine and lysine. The A-band, which is thought to act as a protein-ruler, contains a mixture of immunoglobulin and fibronectin repeats, and possesses kinase activity. An N-terminal Z-disc region and a C-terminal M-line region bind to the Z-line and M-line of the sarcomere, respectively, so that a single titin molecule spans half the length of a sarcomere. Titin also contains binding sites for muscle associated proteins so it serves as an adhesion template for the assembly of contractile machinery in muscle cells. It has also been identified as a structural protein for chromosomes. Alternative splicing of this gene results in multiple transcript variants. Considerable variability exists in the I-band, the M-line and the Z-disc regions of titin. Variability in the I-band region contributes to the differences in elasticity of different titin isoforms and, therefore, to the differences in elasticity of different muscle types. Mutations in this gene are associated with familial hypertrophic cardiomyopathy 9, and autoantibodies to titin are produced in patients with the autoimmune disease scleroderma. [provided by RefSeq, Feb 2012] |
Target gene [TTN] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1010126 |
chr2 |
10 |
3 |
0 |
13 |
View |
Target gene [TTN] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
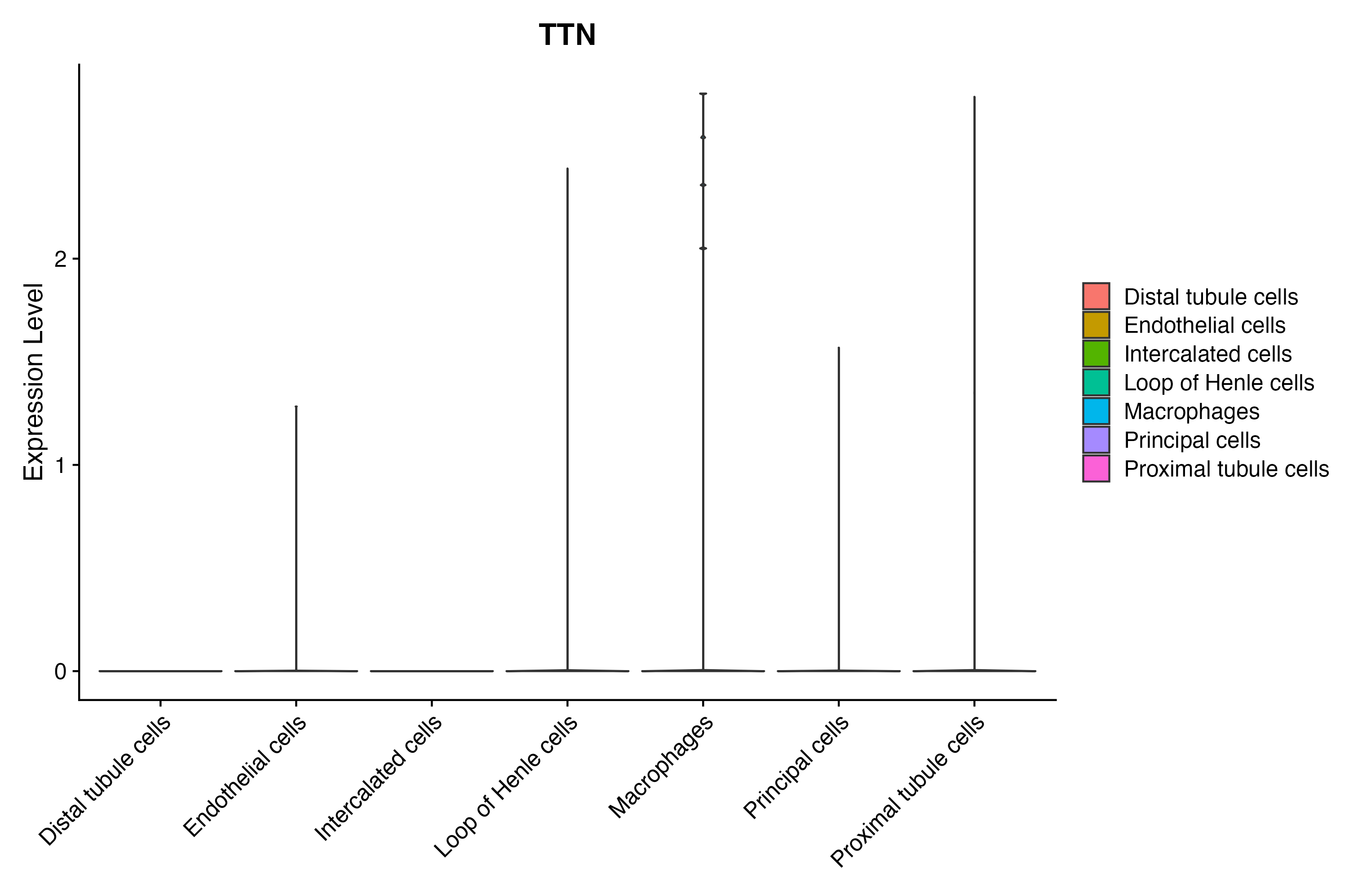
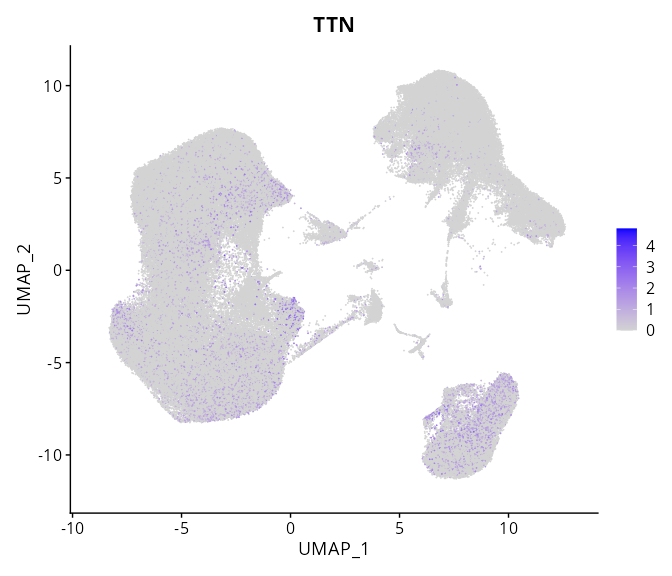
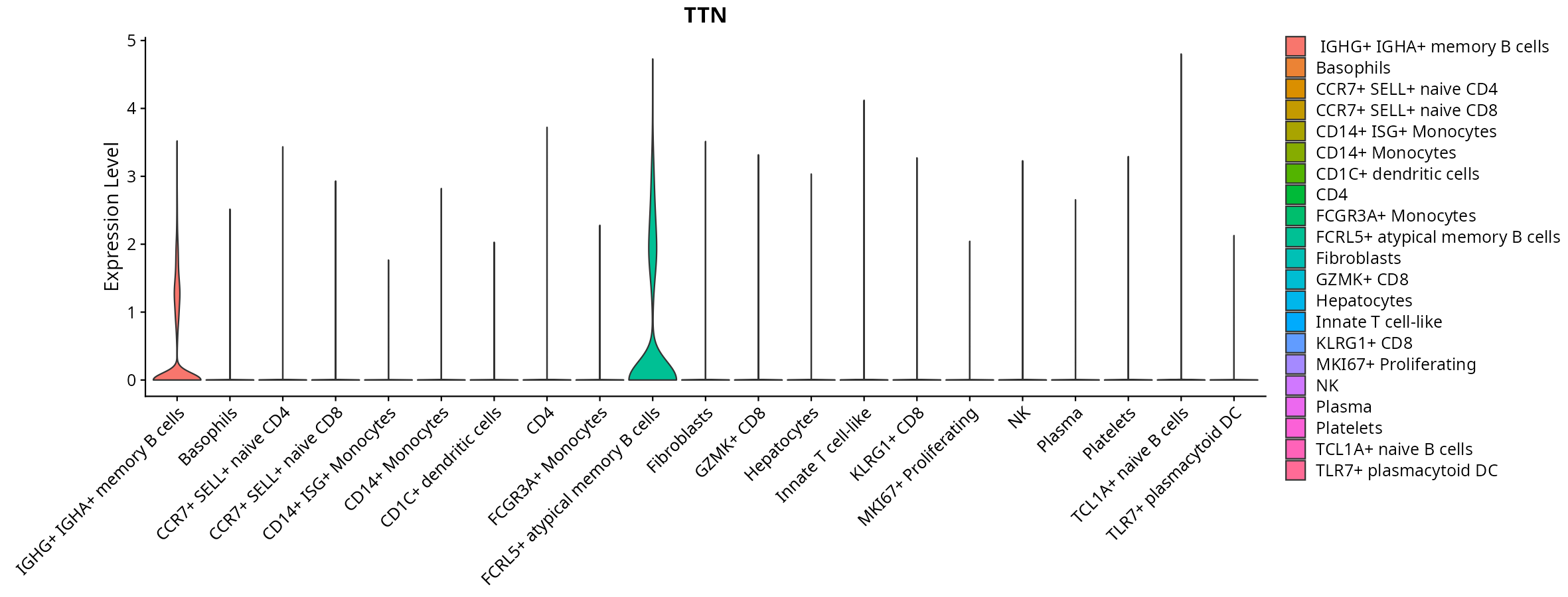
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE199850 - Gene expression in cell subsets
|
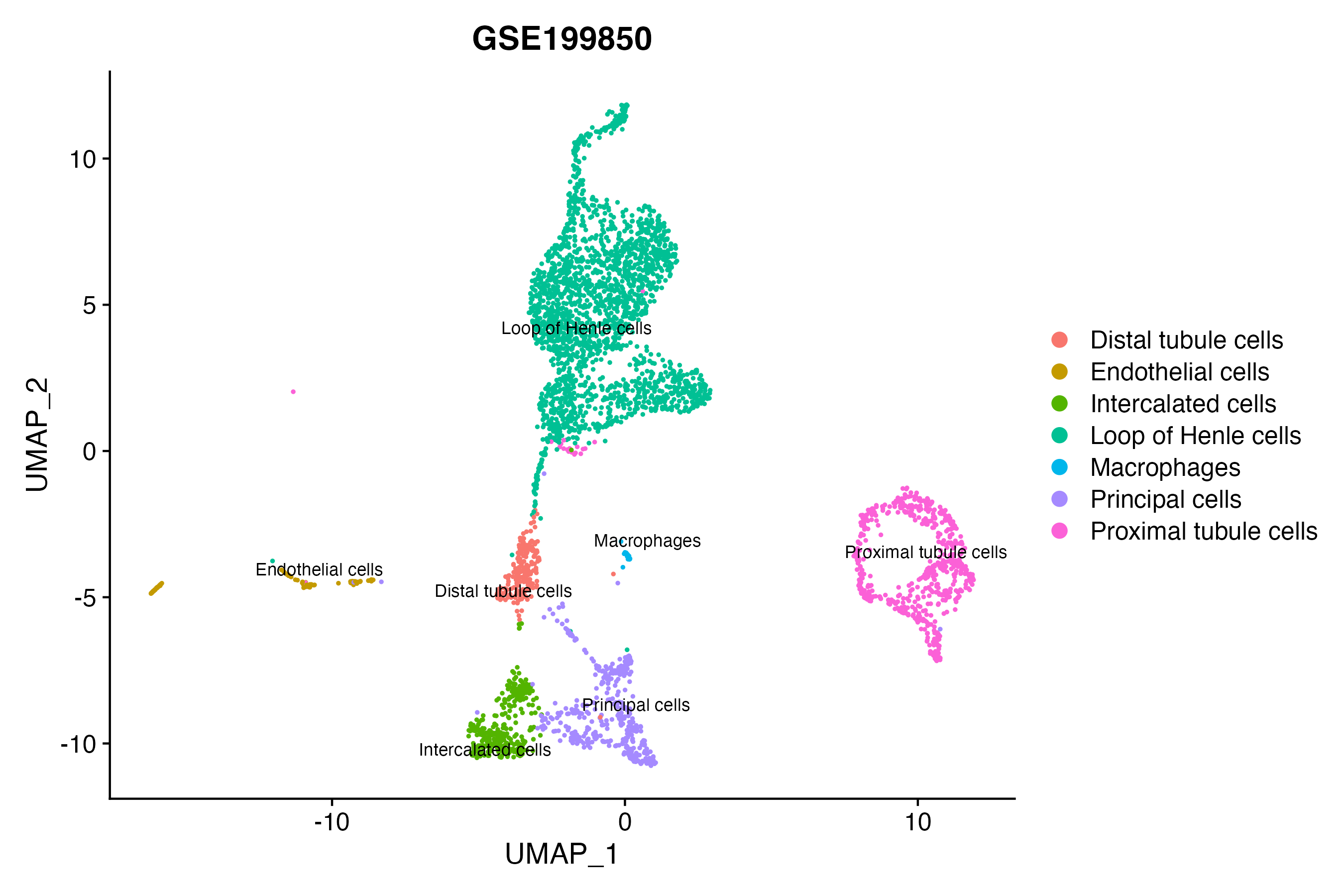
|
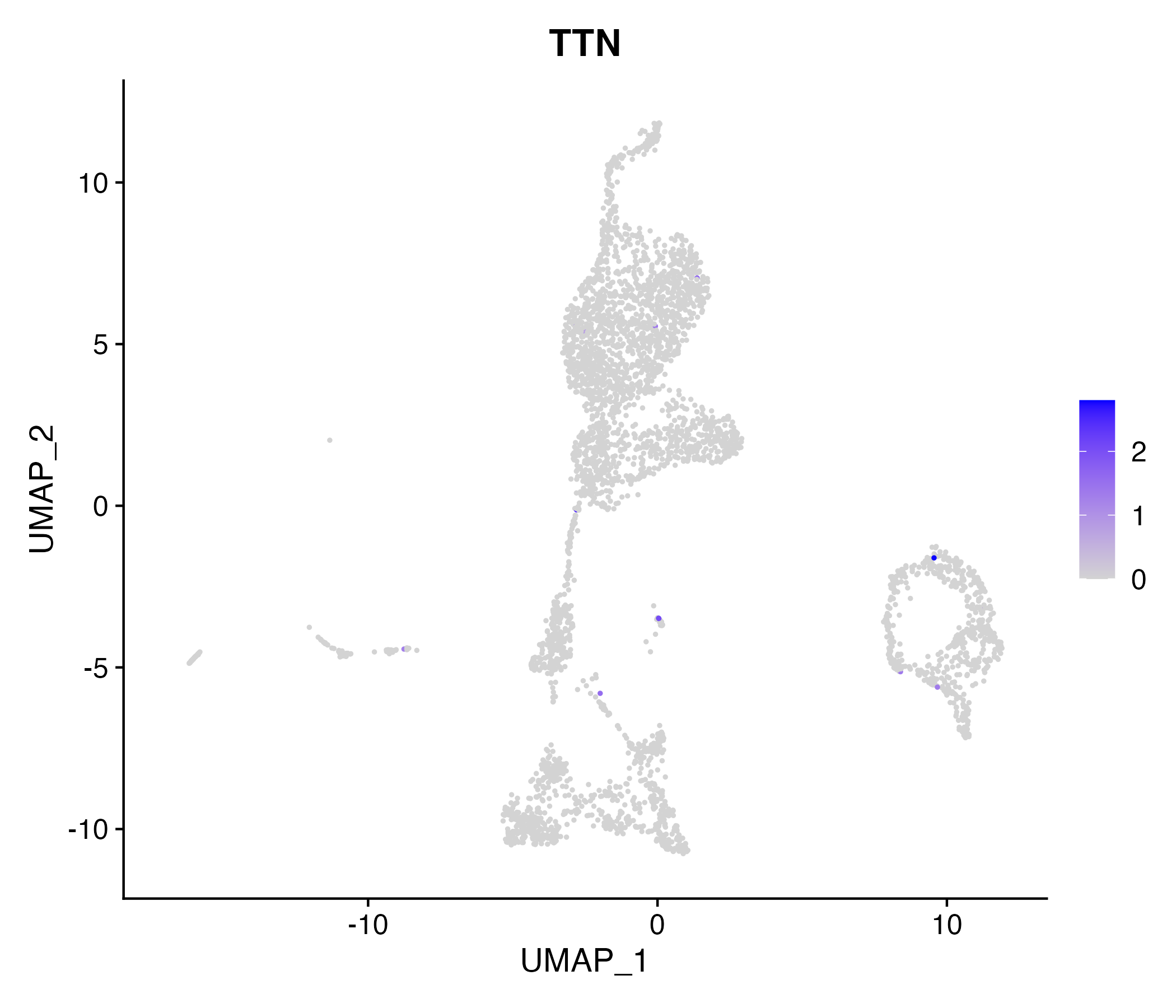
|
|
|
|
> Dataset: GSE247322 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information