Gene Information
|
Gene Name
|
TXNRD3 |
|
Gene ID
|
114112
|
|
Gene Full Name
|
thioredoxin reductase 3 |
|
Gene Alias
|
TGR|TR2|TR2IT1|TRXR3|TXNR3|TXNRD3IT1|TXNRD3NB|TXNRD3NT1 |
|
Transcripts
|
ENSG00000197763
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Cytosol; 
|
|
Membrane Info
|
Enzymes, Metabolic proteins, Predicted intracellular proteins |
|
Uniport_ID
|
Q86VQ6
|
|
HGNC ID
|
HGNC:20667
|
|
OMIM ID
|
606235 |
|
Summary
|
The protein encoded by this gene belongs to the pyridine nucleotide-disulfide oxidoreductase family, and is a member of the thioredoxin (Trx) system. Three thioredoxin reductase (TrxR) isozymes are found in mammals. TrxRs are selenocysteine-containing flavoenzymes, which reduce thioredoxins, as well as other substrates, and play a key role in redox homoeostasis. This gene encodes the third TrxR, which unlike the other two isozymes, contains an additional N-terminal glutaredoxin (Grx) domain, and shows highest expression in testis. The Grx domain allows this isozyme to participate in both Trx and glutathione systems. It functions as a homodimer containing FAD, and selenocysteine (Sec) at the active site. Sec is encoded by UGA codon that normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, the Sec insertion sequence (SECIS) element, which is necessary for the recognition of UGA as a Sec codon rather than as a stop signal. Alternatively spliced transcript variants have been found for this gene. Experimental evidence suggests the use of a non-AUG (CUG) codon as a translation initiation codon (PMID:20018845). [provided by RefSeq, Aug 2017] |
Target gene [TXNRD3] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1005660 |
chr3 |
46 |
2 |
1 |
49 |
View |
Target gene [TXNRD3] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - TXNRD3 peak across samples
|
Peak Plot
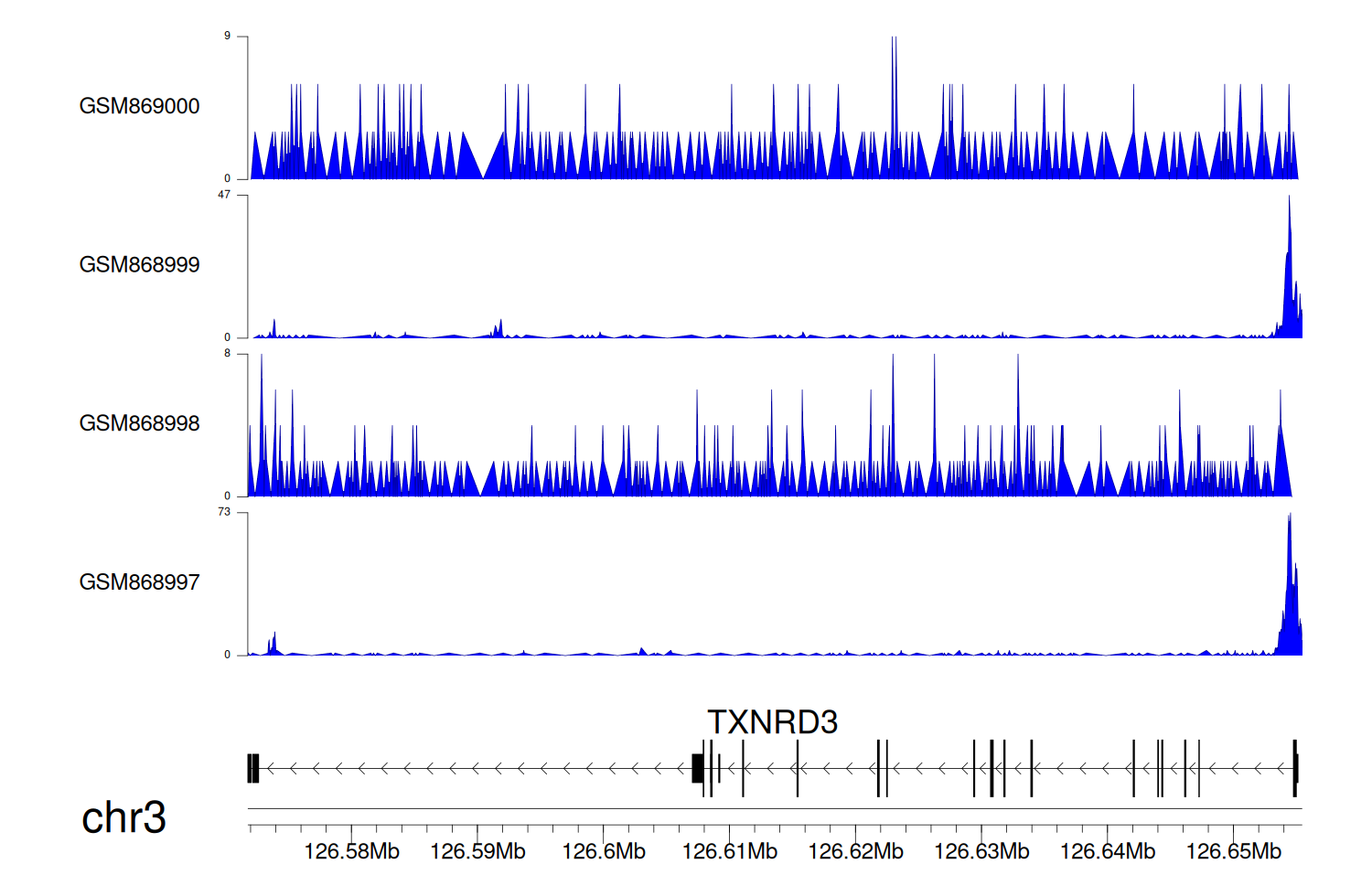
|
> Dataset: GSE68402 - TXNRD3 peak across samples
|
Peak Plot
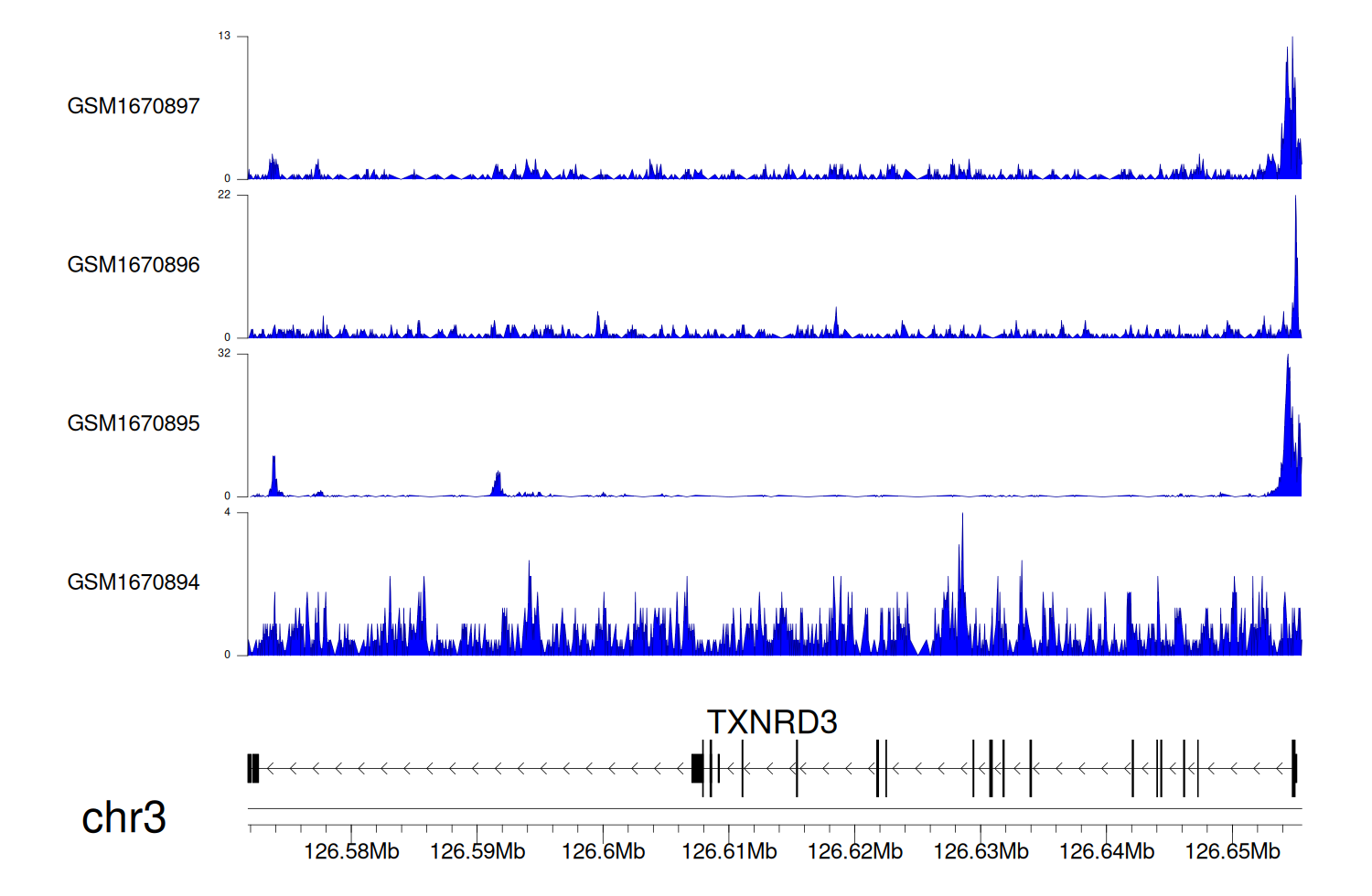
|
> Dataset: GSE270130 - TXNRD3 peak across samples
|
Peak Plot
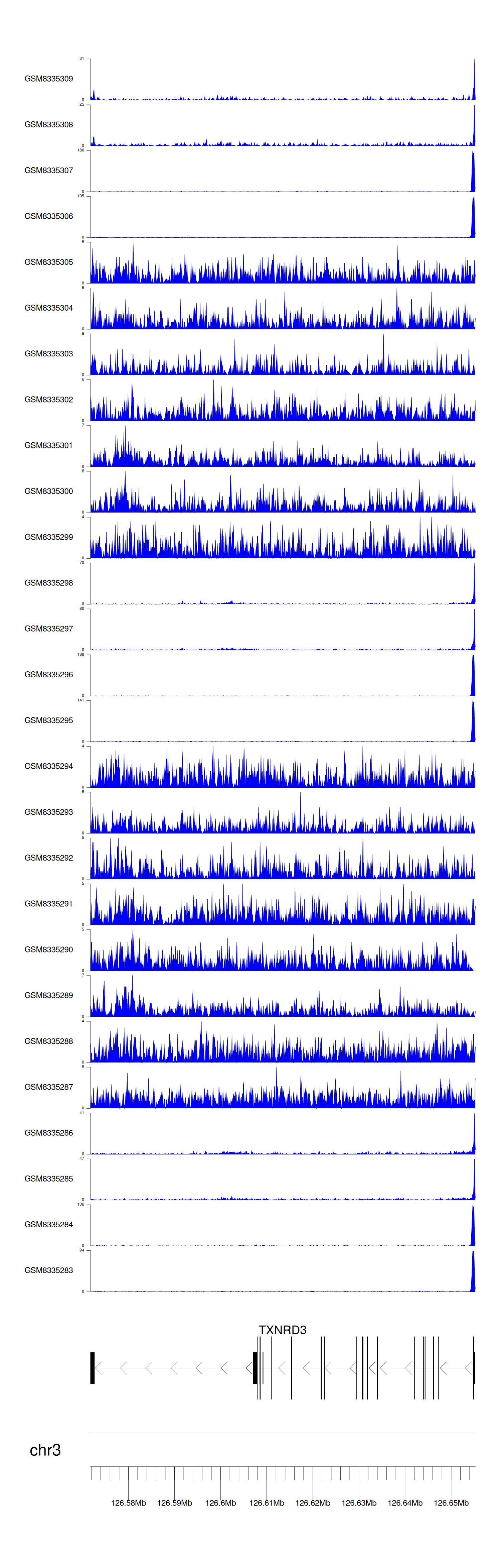
|
> Dataset: GSE131257 - TXNRD3 peak across samples
|
Peak Plot
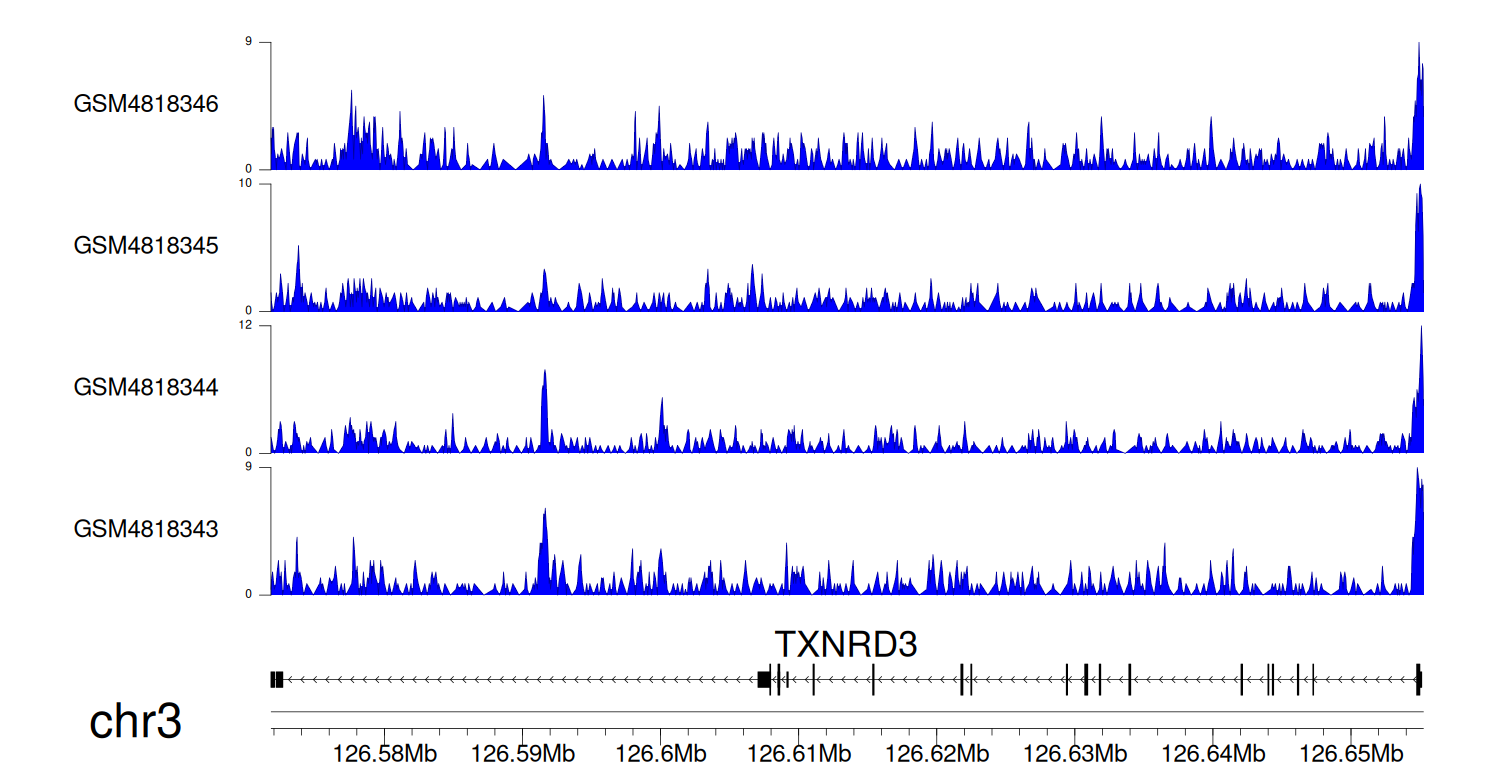
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information