Gene Information
|
Gene Name
|
AKR1C2 |
|
Gene ID
|
1646
|
|
Gene Full Name
|
aldo-keto reductase family 1 member C2 |
|
Gene Alias
|
AKR1C-pseudo|BABP|DD|DD-2|DD/BABP|DD2|DDH2|HAKRD|HBAB|MCDR2|SRXY8|TDD |
|
Transcripts
|
ENSG00000151632
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm, Cytosol;Nucleoli fibrillar center, Endoplasmic reticulum; 
|
|
Membrane Info
|
Disease related genes, Enzymes, FDA approved drug targets, Human disease related genes, Metabolic proteins, Predicted intracellular proteins |
|
Uniport_ID
|
P52895
|
|
HGNC ID
|
HGNC:385
|
|
OMIM ID
|
600450 |
|
Summary
|
This gene encodes a member of the aldo/keto reductase superfamily, which consists of more than 40 known enzymes and proteins. These enzymes catalyze the conversion of aldehydes and ketones to their corresponding alcohols using NADH and/or NADPH as cofactors. The enzymes display overlapping but distinct substrate specificity. This enzyme binds bile acid with high affinity, and shows minimal 3-alpha-hydroxysteroid dehydrogenase activity. This gene shares high sequence identity with three other gene members and is clustered with those three genes at chromosome 10p15-p14. Three transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Dec 2011] |
Target gene [AKR1C2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1004665 |
chr10 |
50 |
13 |
9 |
72 |
View |
| 1022889 |
chr10 |
50 |
13 |
9 |
72 |
View |
Target gene [AKR1C2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
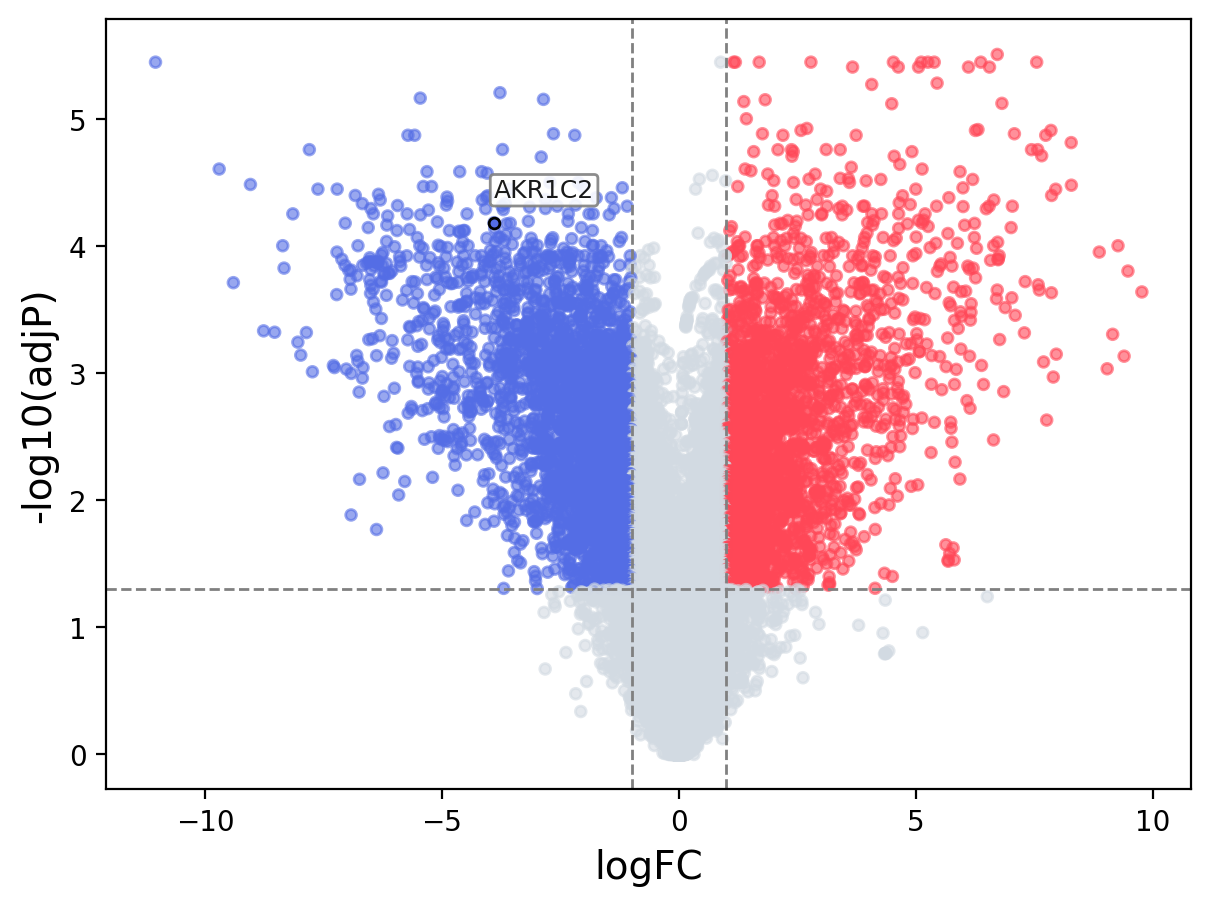
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - AKR1C2 expression across samples
|
Volcano Plot
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information