Gene Information
|
Gene Name
|
BTRC |
|
Gene ID
|
8945
|
|
Gene Full Name
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
|
Gene Alias
|
BETA-TRCP|FBW1A|FBXW1|FBXW1A|FWD1|bTrCP|bTrCP1|betaTrCP |
|
Transcripts
|
ENSG00000166167
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Plasma membrane, Cytosol; 
|
|
Membrane Info
|
Predicted intracellular proteins |
|
Uniport_ID
|
Q9Y297
|
|
HGNC ID
|
HGNC:1144
|
|
OMIM ID
|
603482 |
|
Summary
|
This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into 3 classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbws class; in addition to an F-box, this protein contains multiple WD-40 repeats. The encoded protein mediates degradation of CD4 via its interaction with HIV-1 Vpu. It has also been shown to ubiquitinate phosphorylated NFKBIA (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), targeting it for degradation and thus activating nuclear factor kappa-B. Alternatively spliced transcript variants have been described. A related pseudogene exists in chromosome 6. [provided by RefSeq, Mar 2012] |
Target gene [BTRC] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1013723 |
chr10 |
2 |
2 |
0 |
4 |
View |
| 1022937 |
chr10 |
2 |
2 |
0 |
4 |
View |
Target gene [BTRC] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - BTRC peak across samples
|
Peak Plot
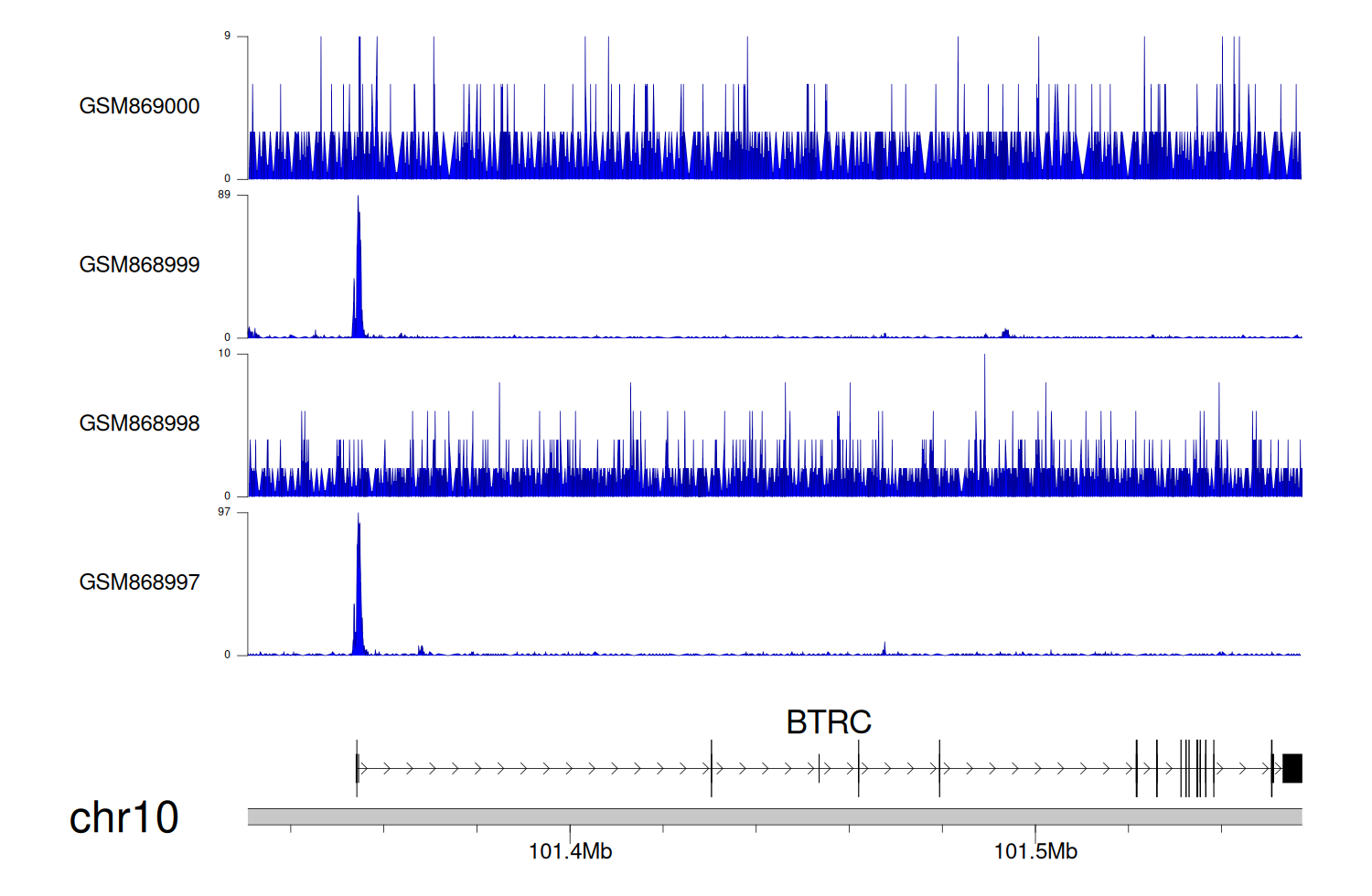
|
> Dataset: GSE68402 - BTRC peak across samples
|
Peak Plot
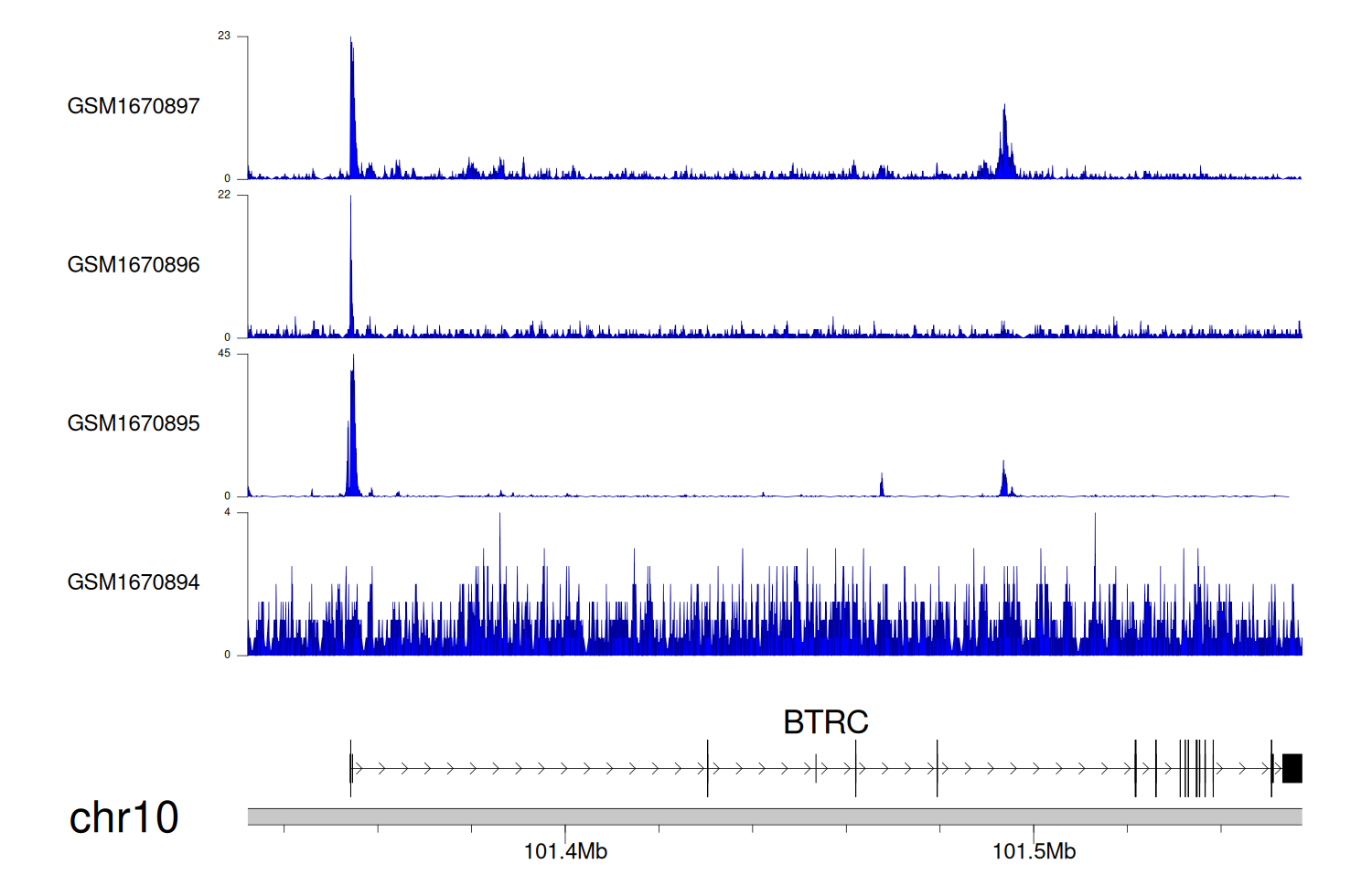
|
> Dataset: GSE270130 - BTRC peak across samples
|
Peak Plot
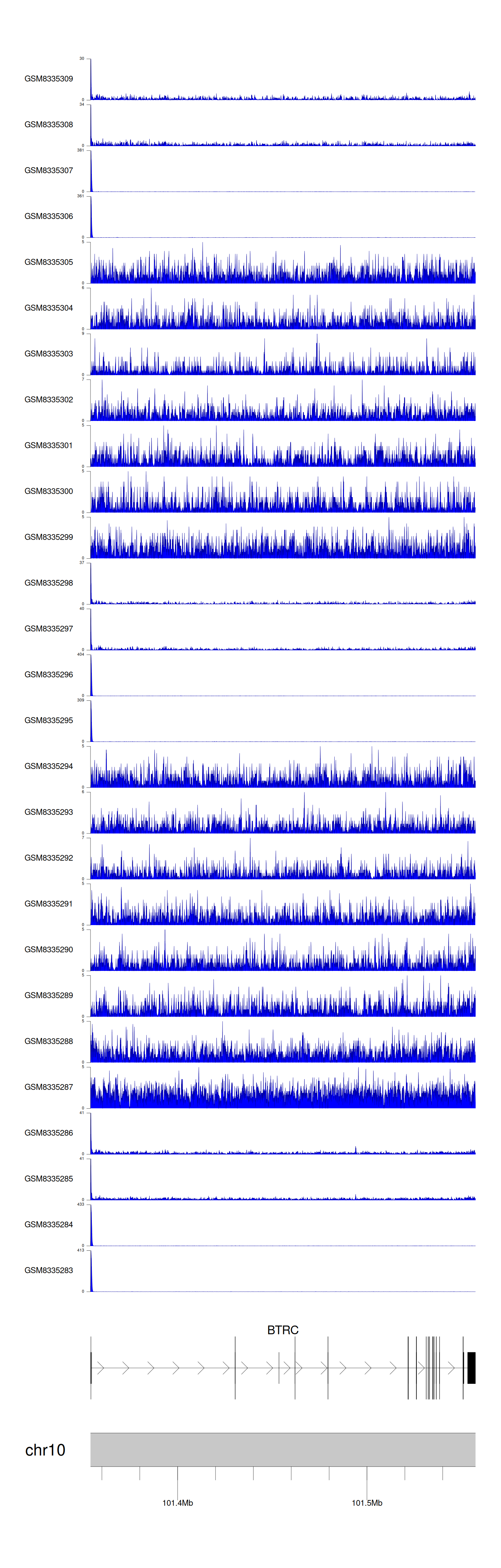
|
> Dataset: GSE131257 - BTRC peak across samples
|
Peak Plot
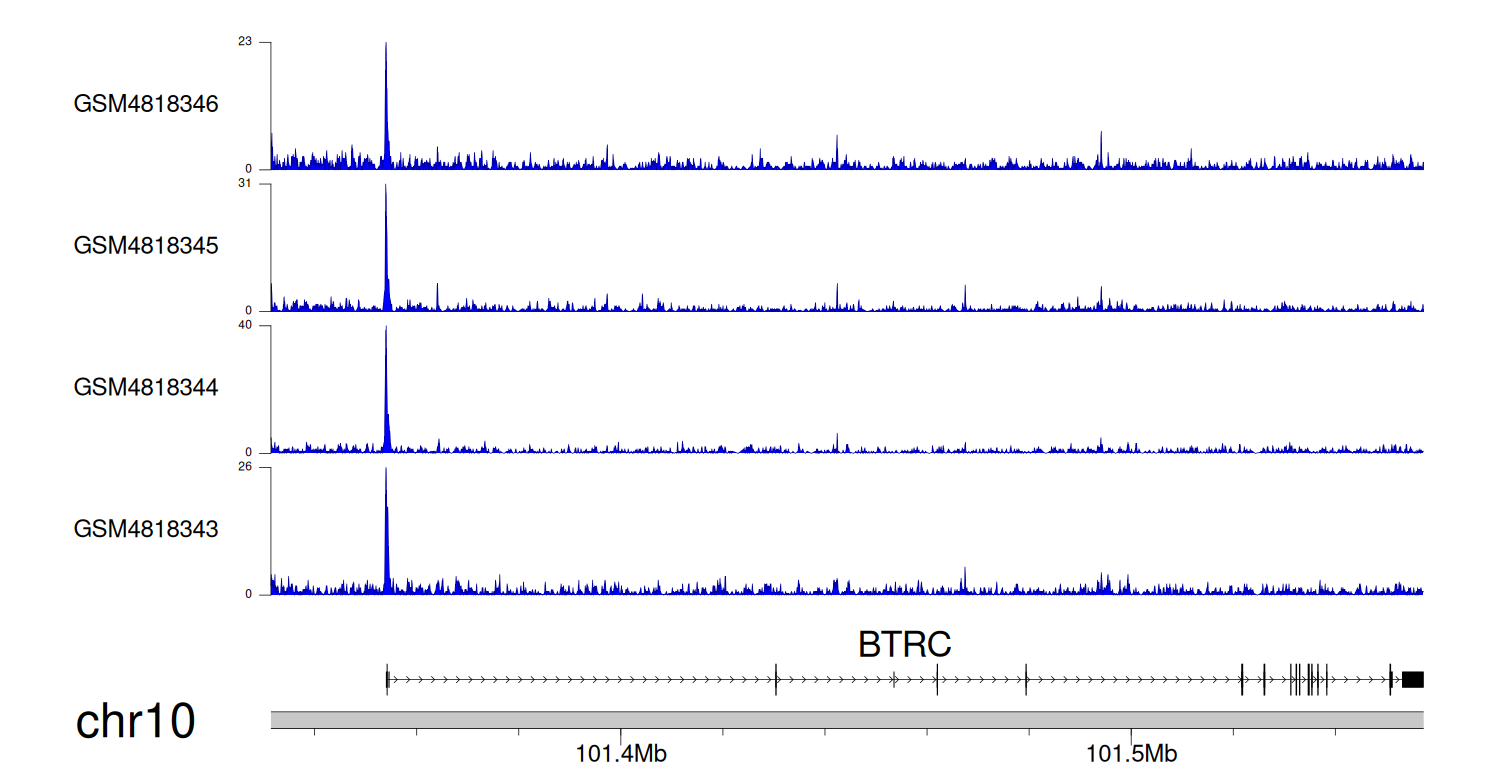
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information