Gene Information
|
Gene Name
|
CNTNAP2 |
|
Gene ID
|
26047
|
|
Gene Full Name
|
contactin associated protein 2 |
|
Gene Alias
|
AUTS15|CASPR2|CDFE|NRXN4|PTHSL1 |
|
Transcripts
|
ENSG00000174469
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
Q9UHC6
|
|
HGNC ID
|
HGNC:13830
|
|
OMIM ID
|
604569 |
|
Summary
|
This gene encodes a member of the neurexin family which functions in the vertebrate nervous system as cell adhesion molecules and receptors. This protein, like other neurexin proteins, contains epidermal growth factor repeats and laminin G domains. In addition, it includes an F5/8 type C domain, discoidin/neuropilin- and fibrinogen-like domains, thrombospondin N-terminal-like domains and a putative PDZ binding site. This protein is localized at the juxtaparanodes of myelinated axons, and mediates interactions between neurons and glia during nervous system development and is also involved in localization of potassium channels within differentiating axons. This gene encompasses almost 1.5% of chromosome 7 and is one of the largest genes in the human genome. It is directly bound and regulated by forkhead box protein P2, a transcription factor related to speech and language development. This gene has been implicated in multiple neurodevelopmental disorders, including Gilles de la Tourette syndrome, schizophrenia, epilepsy, autism, ADHD and intellectual disability. [provided by RefSeq, Jul 2017] |
Target gene [CNTNAP2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1005466 |
chr7 |
0 |
0 |
54 |
54 |
View |
| 1005950 |
chr7 |
14 |
26 |
5 |
45 |
View |
| 1008878 |
chr7 |
5 |
13 |
1 |
19 |
View |
| 1013389 |
chr7 |
2 |
3 |
0 |
5 |
View |
| 1013621 |
chr7 |
5 |
0 |
0 |
5 |
View |
| 1014875 |
chr7 |
3 |
0 |
0 |
3 |
View |
| 1016825 |
chr7 |
1 |
0 |
0 |
1 |
View |
| 1017606 |
chr7 |
0 |
0 |
0 |
0 |
View |
| 1017730 |
chr7 |
0 |
0 |
0 |
0 |
View |
| 1023068 |
chr7 |
0 |
0 |
0 |
0 |
View |
| 1023069 |
chr7 |
0 |
0 |
0 |
0 |
View |
| 1041559 |
chr7 |
3 |
3 |
3 |
9 |
View |
| 1043284 |
chr7 |
2 |
0 |
0 |
2 |
View |
| 1043285 |
chr7 |
21 |
24 |
66 |
111 |
View |
Target gene [CNTNAP2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
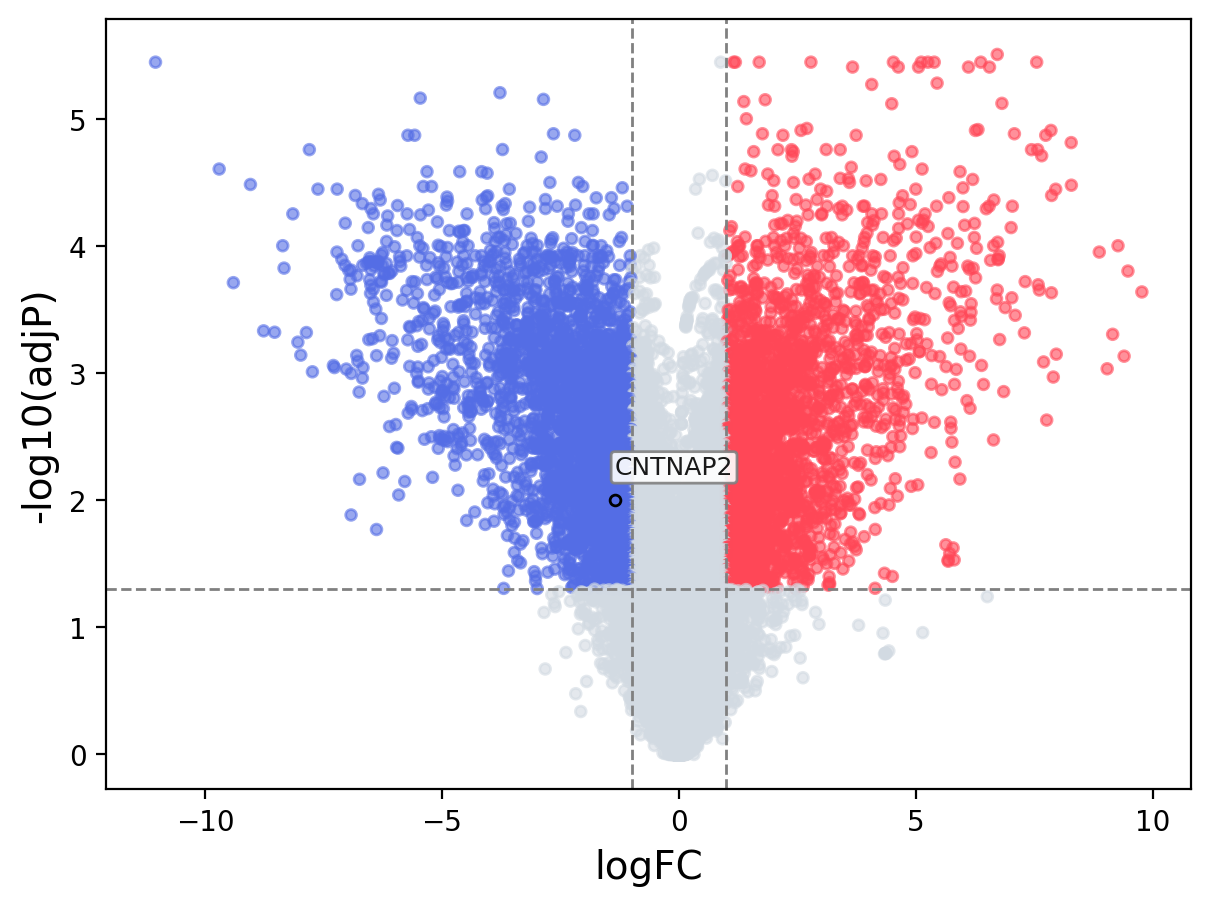
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - CNTNAP2 expression across samples
|
Volcano Plot
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information