Gene Information
|
Gene Name
|
GATM |
|
Gene ID
|
2628
|
|
Gene Full Name
|
glycine amidinotransferase |
|
Gene Alias
|
AGAT|AT|CCDS3|FRTS|FRTS1|RFS |
|
Transcripts
|
ENSG00000171766
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Mitochondria; 
|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Plasma proteins, Potential drug targets, Predicted intracellular proteins |
|
Uniport_ID
|
P50440
|
|
HGNC ID
|
HGNC:4175
|
|
OMIM ID
|
602360 |
|
Summary
|
This gene encodes a mitochondrial enzyme that belongs to the amidinotransferase family. This enzyme is involved in creatine biosynthesis, whereby it catalyzes the transfer of a guanido group from L-arginine to glycine, resulting in guanidinoacetic acid, the immediate precursor of creatine. Mutations in this gene cause arginine:glycine amidinotransferase deficiency, an inborn error of creatine synthesis characterized by cognitive disability, language impairment, and behavioral disorders. [provided by RefSeq, Jul 2008] |
Target gene [GATM] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1004524 |
chr15 |
27 |
42 |
9 |
78 |
View |
| 1008107 |
chr15 |
18 |
4 |
0 |
22 |
View |
| 1008108 |
chr15 |
18 |
4 |
0 |
22 |
View |
| 1011161 |
chr15 |
4 |
1 |
5 |
10 |
View |
| 1039441 |
chr15 |
9 |
0 |
0 |
9 |
View |
Target gene [GATM] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
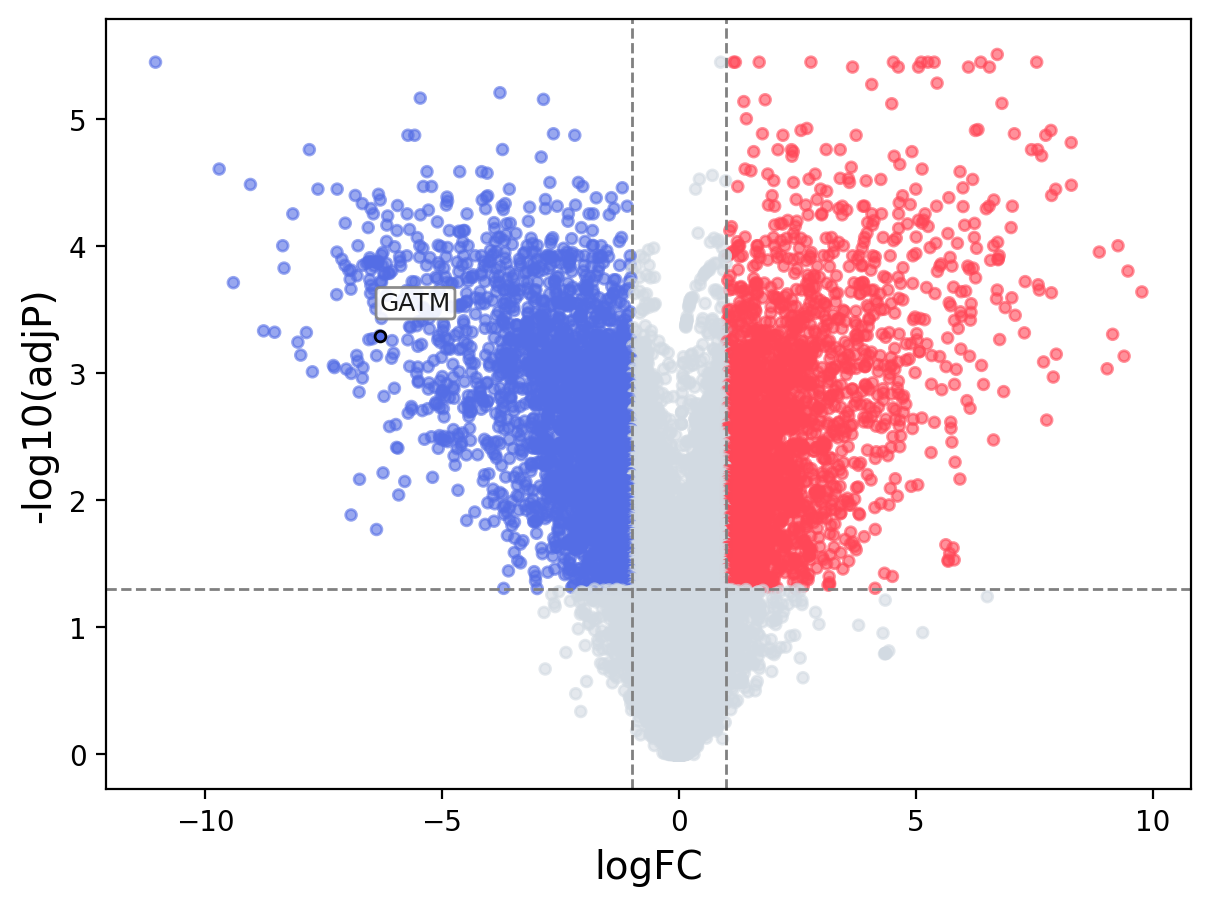
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - GATM expression across samples
|
Volcano Plot
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information