Gene Information
|
Gene Name
|
HTT |
|
Gene ID
|
3064
|
|
Gene Full Name
|
huntingtin |
|
Gene Alias
|
HD|IT15|LOMARS |
|
Transcripts
|
ENSG00000197386
|
|
Virus
|
HTLV1 |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Cytosol; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P42858
|
|
HGNC ID
|
HGNC:4851
|
|
OMIM ID
|
613004 |
|
Summary
|
Huntingtin is a disease gene linked to Huntington's disease, a neurodegenerative disorder characterized by loss of striatal neurons. This is thought to be caused by an expanded, unstable trinucleotide repeat in the huntingtin gene, which translates as a polyglutamine repeat in the protein product. A fairly broad range of trinucleotide repeats (9-35) has been identified in normal controls, and repeat numbers in excess of 40 have been described as pathological. The huntingtin locus is large, spanning 180 kb and consisting of 67 exons. The huntingtin gene is widely expressed and is required for normal development. It is expressed as 2 alternatively polyadenylated forms displaying different relative abundance in various fetal and adult tissues. The larger transcript is approximately 13.7 kb and is expressed predominantly in adult and fetal brain whereas the smaller transcript of approximately 10.3 kb is more widely expressed. The genetic defect leading to Huntington's disease may not necessarily eliminate transcription, but may confer a new property on the mRNA or alter the function of the protein. One candidate is the huntingtin-associated protein-1, highly expressed in brain, which has increased affinity for huntingtin protein with expanded polyglutamine repeats. This gene contains an upstream open reading frame in the 5' UTR that inhibits expression of the huntingtin gene product through translational repression. [provided by RefSeq, Jul 2016] |
Target gene [HTT] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 6007909 |
chr4 |
10 |
40 |
0 |
50 |
View |
| 6011169 |
chr4 |
10 |
16 |
1 |
27 |
View |
Target gene [HTT] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE94732
|
Chip-seq |
24 |
Illumina NextSeq 500 (Homo sapiens);illumina Genome Analyzer IIx (Homo sapiens) |
|
GSE52244
|
Expression array |
15 |
[HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [probe set (exon) version] |
|
GSE10789
|
Expression array |
6 |
NCI/ATC Hs-OperonV3 |
|
GSE224047
|
RNA-seq |
10 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE189602
|
Methylation profiling (Array) |
4 |
Infinium MethylationEPIC |
|
GSE136189
|
Methylation profiling (Array) |
40 |
Illumina HumanMethylation450 BeadChip (HumanMethylation450_15017482);Illumina Infinium HumanMethylation850 BeadChip |
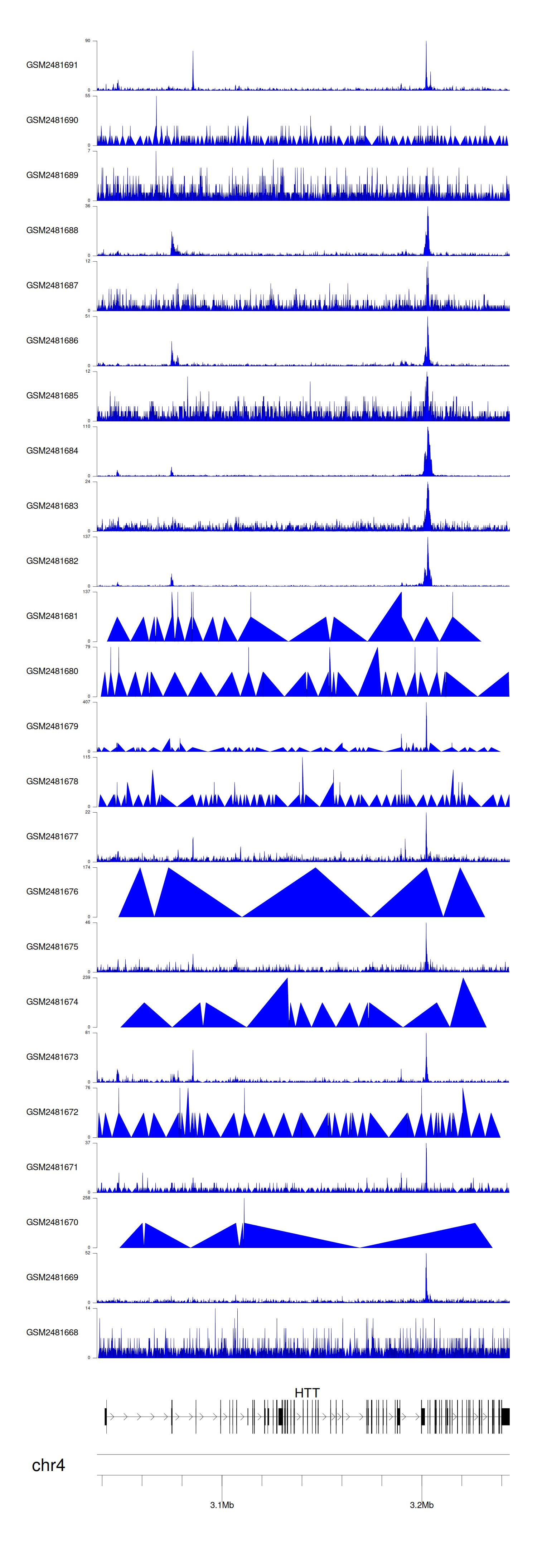
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE94732 - HTT peak across samples
|
Peak Plot
|
|
|
 HTLV1 Target gene Detail Information
HTLV1 Target gene Detail Information