Gene Information
|
Gene Name
|
NCAM1 |
|
Gene ID
|
4684
|
|
Gene Full Name
|
neural cell adhesion molecule 1 |
|
Gene Alias
|
CD56|MSK39|NCAM |
|
Transcripts
|
ENSG00000149294
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Cytosol;Secreted in other tissues; 
|
|
Membrane Info
|
Cancer-related genes, CD markers, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins, Predicted secreted proteins, Transporters |
|
Uniport_ID
|
P13591
|
|
HGNC ID
|
HGNC:7656
|
|
OMIM ID
|
116930 |
|
Summary
|
This gene encodes a cell adhesion protein which is a member of the immunoglobulin superfamily. The encoded protein is involved in cell-to-cell interactions as well as cell-matrix interactions during development and differentiation. The encoded protein plays a role in the development of the nervous system by regulating neurogenesis, neurite outgrowth, and cell migration. This protein is also involved in the expansion of T lymphocytes, B lymphocytes and natural killer (NK) cells which play an important role in immune surveillance. This protein plays a role in signal transduction by interacting with fibroblast growth factor receptors, N-cadherin and other components of the extracellular matrix and by triggering signalling cascades involving FYN-focal adhesion kinase (FAK), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K). One prominent isoform of this gene, cell surface molecule CD56, plays a role in several myeloproliferative disorders such as acute myeloid leukemia and differential expression of this gene is associated with differential disease progression. For example, increased expression of CD56 is correlated with lower survival in acute myeloid leukemia patients whereas increased severity of COVID-19 is correlated with decreased abundance of CD56-expressing NK cells in peripheral blood. Alternative splicing results in multiple transcript variants encoding distinct protein isoforms. [provided by RefSeq, Aug 2020] |
Target gene [NCAM1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1007578 |
chr11 |
7 |
12 |
7 |
26 |
View |
| 1041045 |
chr11 |
14 |
1 |
0 |
15 |
View |
Target gene [NCAM1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - NCAM1 expression across samples
|
Volcano Plot
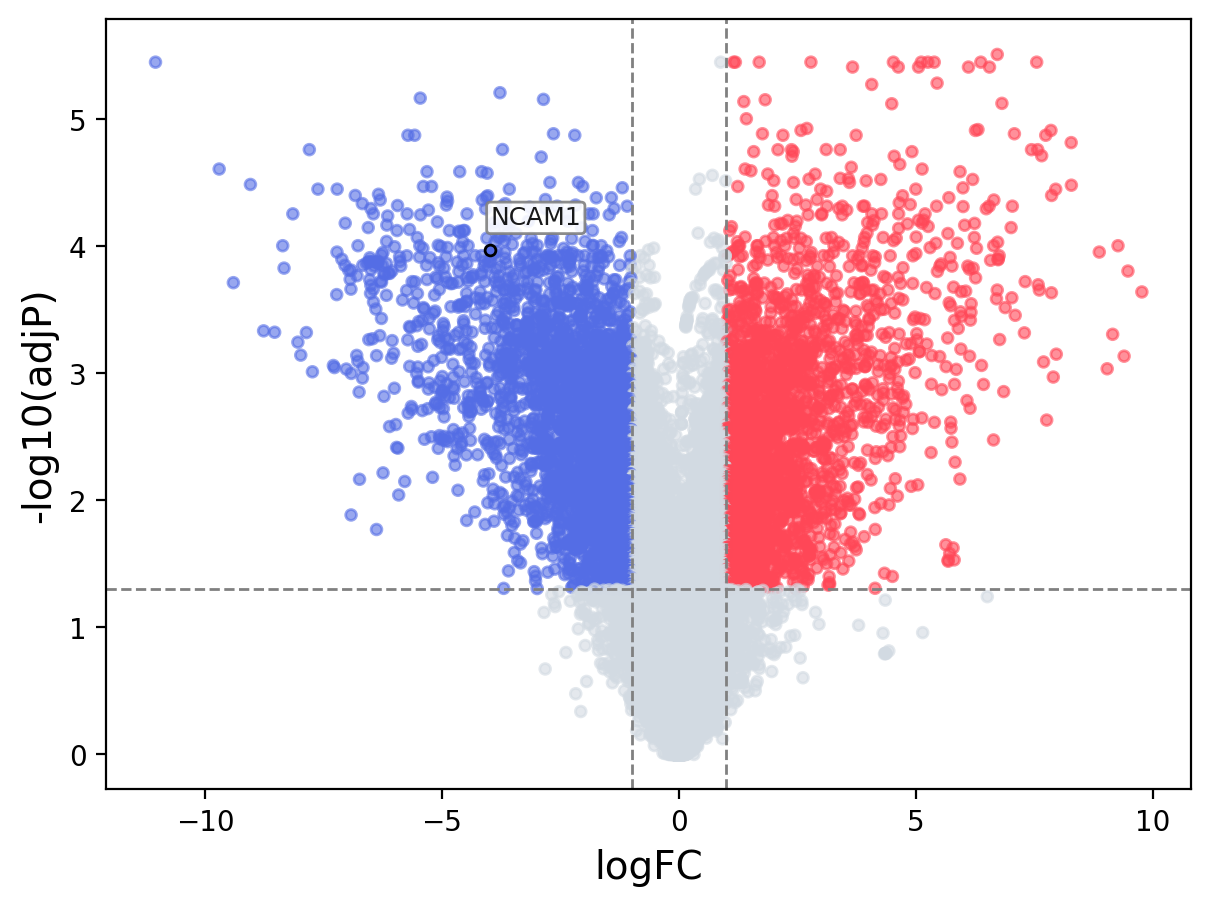
|
Bar Plot
|
> Dataset: GSE94660 - NCAM1 expression across samples
|
Volcano Plot
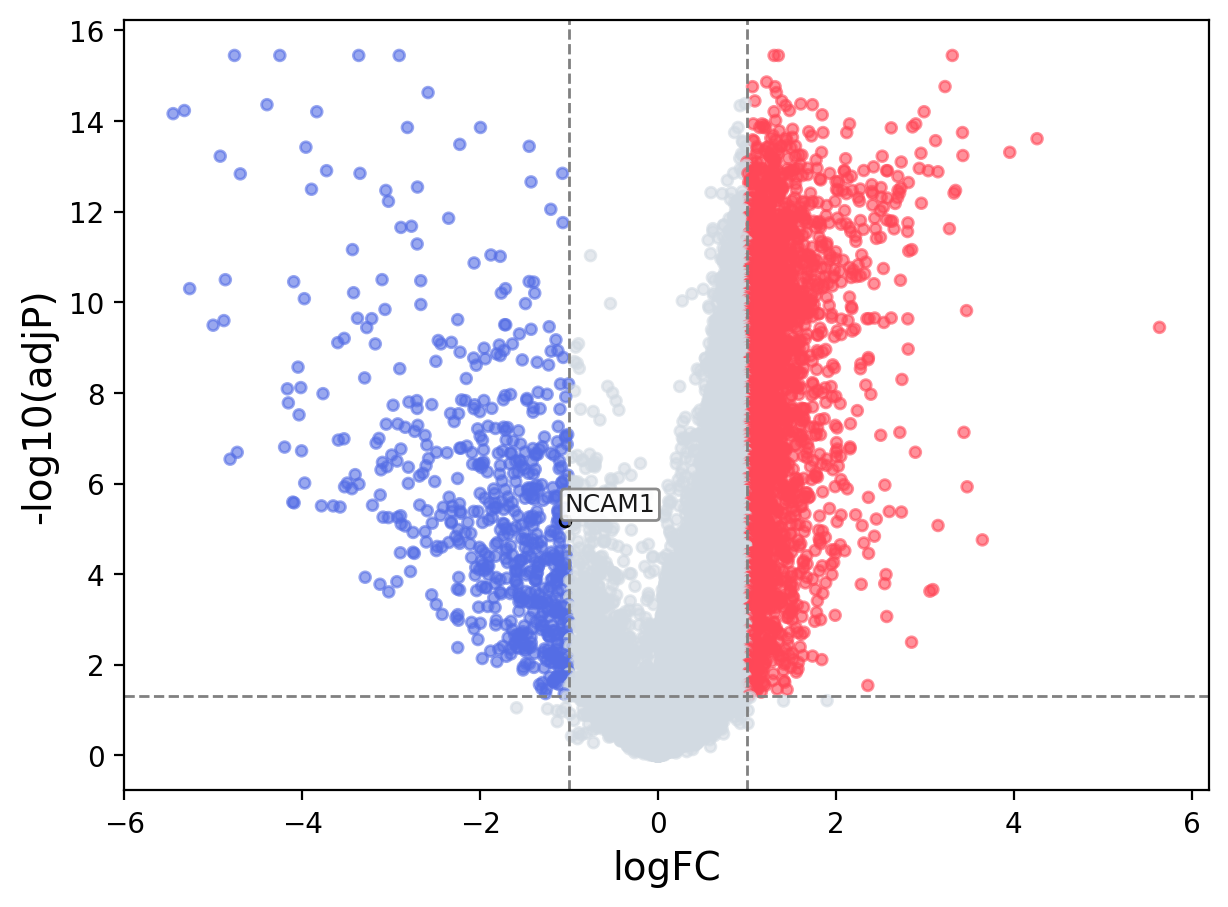
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information