Gene Information
|
Gene Name
|
ODF2 |
|
Gene ID
|
4957
|
|
Gene Full Name
|
outer dense fiber of sperm tails 2 |
|
Gene Alias
|
CT134|ODF2/1|ODF2/2|ODF84 |
|
Transcripts
|
ENSG00000136811
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Primary cilium, Centrosome, Basal body, Connecting piece;Vesicles, Microtubules, Calyx, Mid piece, Principal piece; 
|
|
Membrane Info
|
Predicted intracellular proteins |
|
Uniport_ID
|
Q5BJF6
|
|
HGNC ID
|
HGNC:8114
|
|
OMIM ID
|
602015 |
|
Summary
|
The outer dense fibers are cytoskeletal structures that surround the axoneme in the middle piece and principal piece of the sperm tail. The fibers function in maintaining the elastic structure and recoil of the sperm tail as well as in protecting the tail from shear forces during epididymal transport and ejaculation. Defects in the outer dense fibers lead to abnormal sperm morphology and infertility. This gene encodes one of the major outer dense fiber proteins. Alternative splicing results in multiple transcript variants. The longer transcripts, also known as 'Cenexins', encode proteins with a C-terminal extension that are differentially targeted to somatic centrioles and thought to be crucial for the formation of microtubule organizing centers. [provided by RefSeq, Oct 2010] |
Target gene [ODF2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1005417 |
chr9 |
23 |
23 |
9 |
55 |
View |
Target gene [ODF2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - ODF2 peak across samples
|
Peak Plot
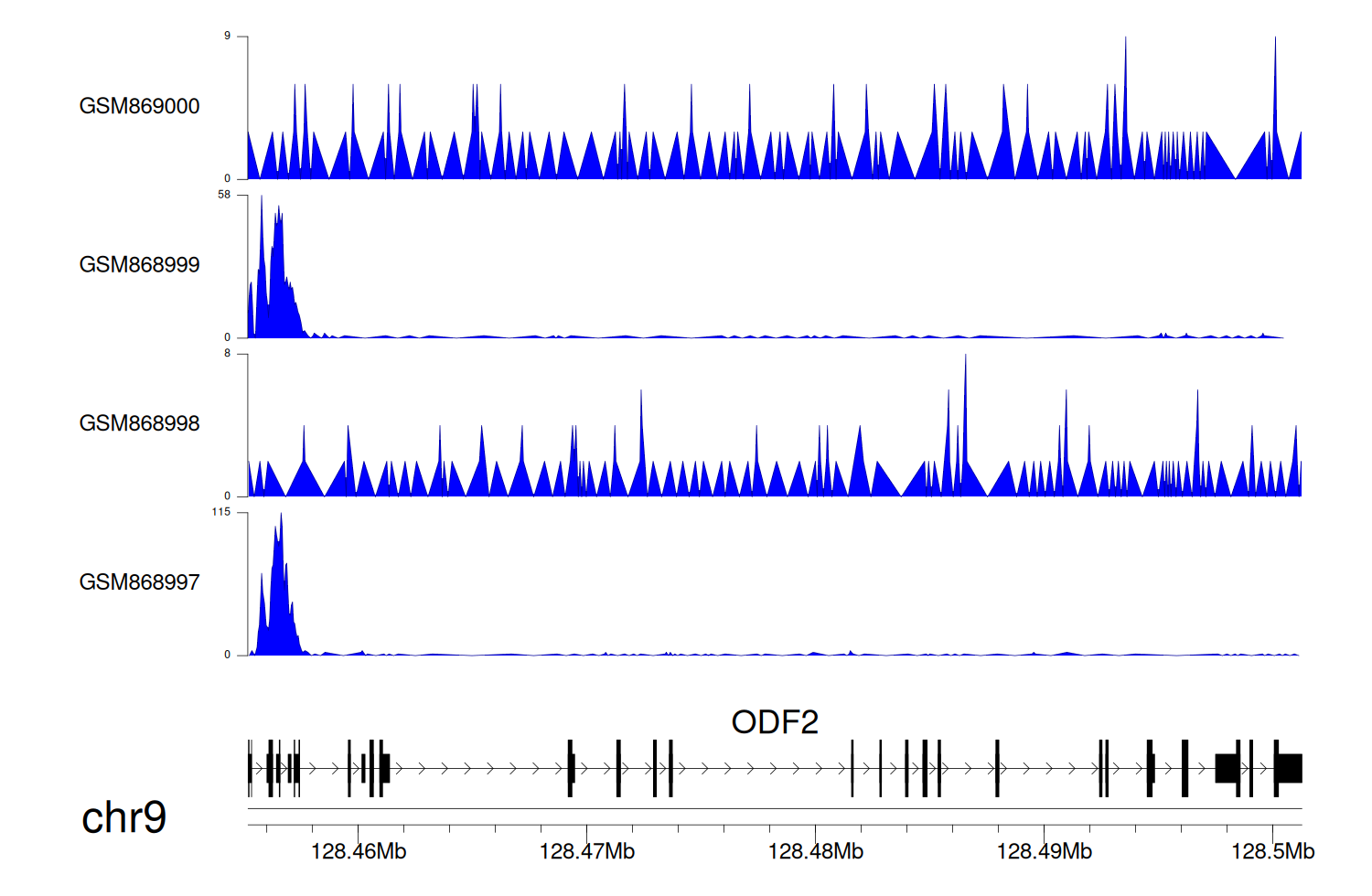
|
> Dataset: GSE68402 - ODF2 peak across samples
|
Peak Plot
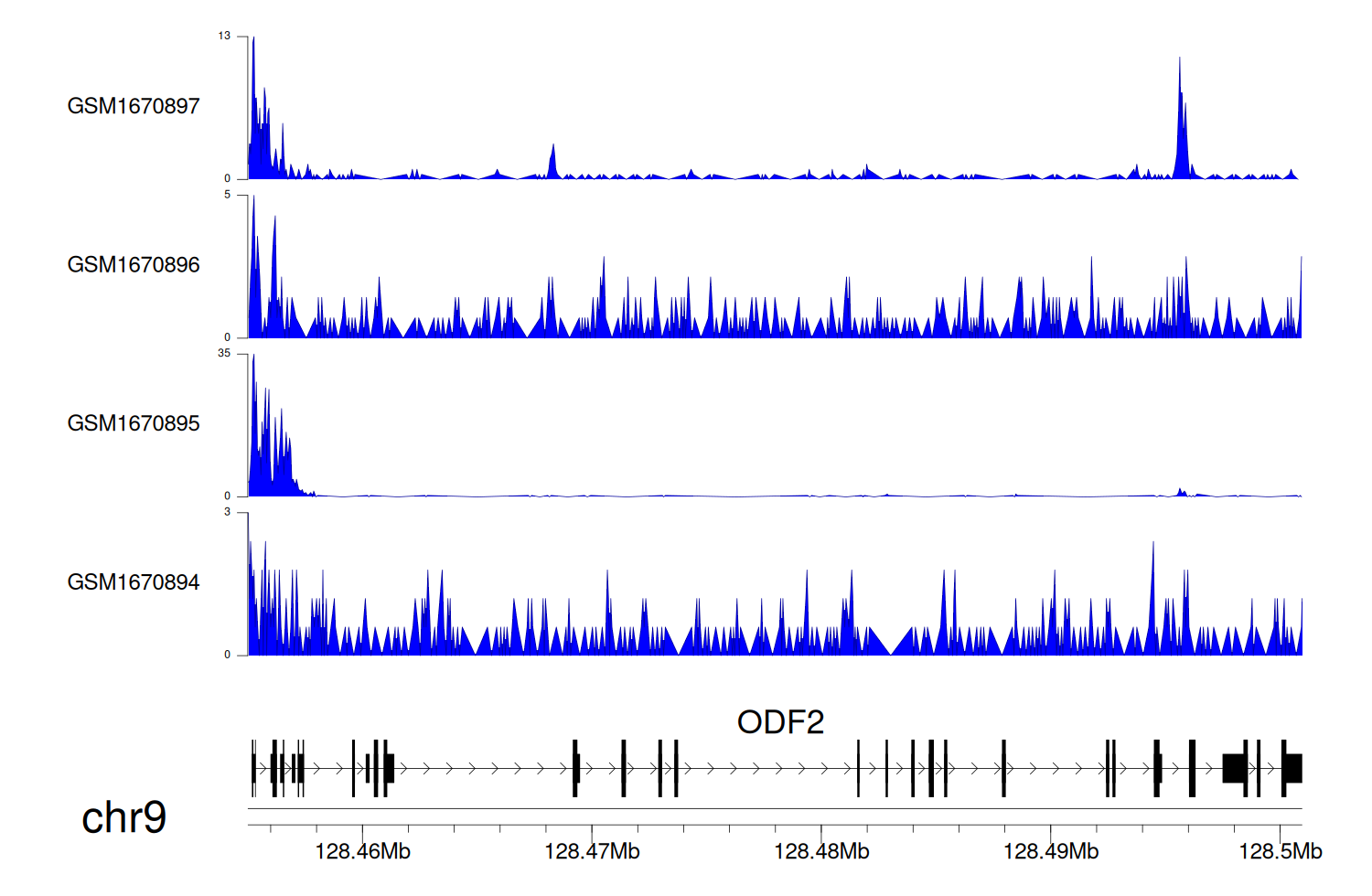
|
> Dataset: GSE270130 - ODF2 peak across samples
|
Peak Plot
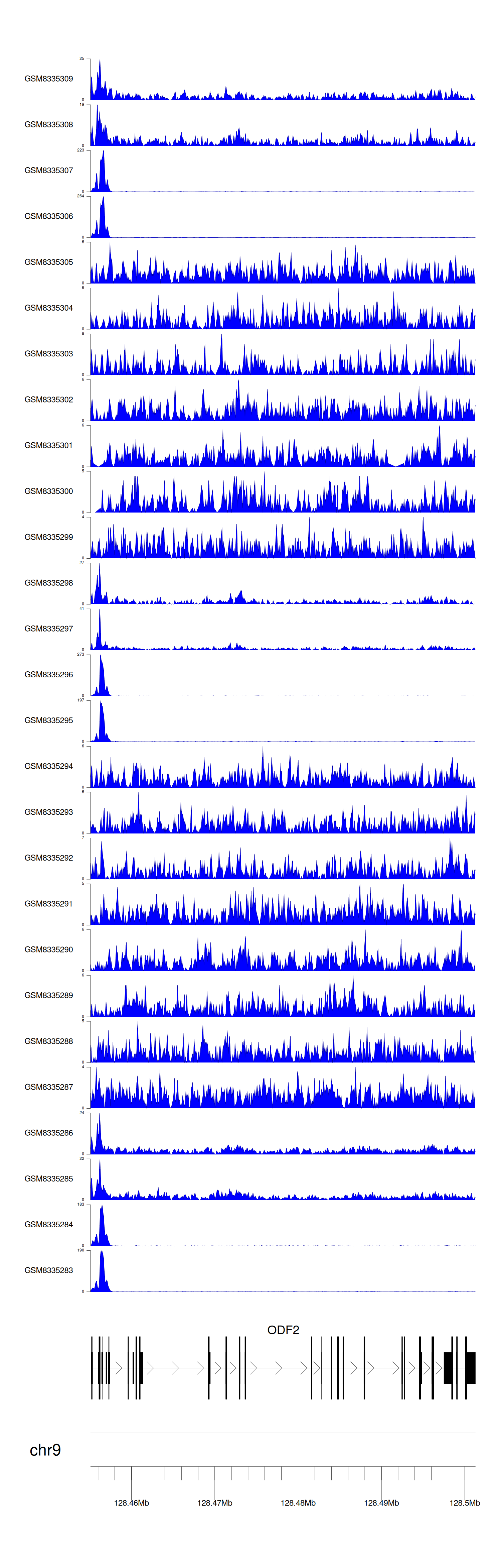
|
> Dataset: GSE131257 - ODF2 peak across samples
|
Peak Plot
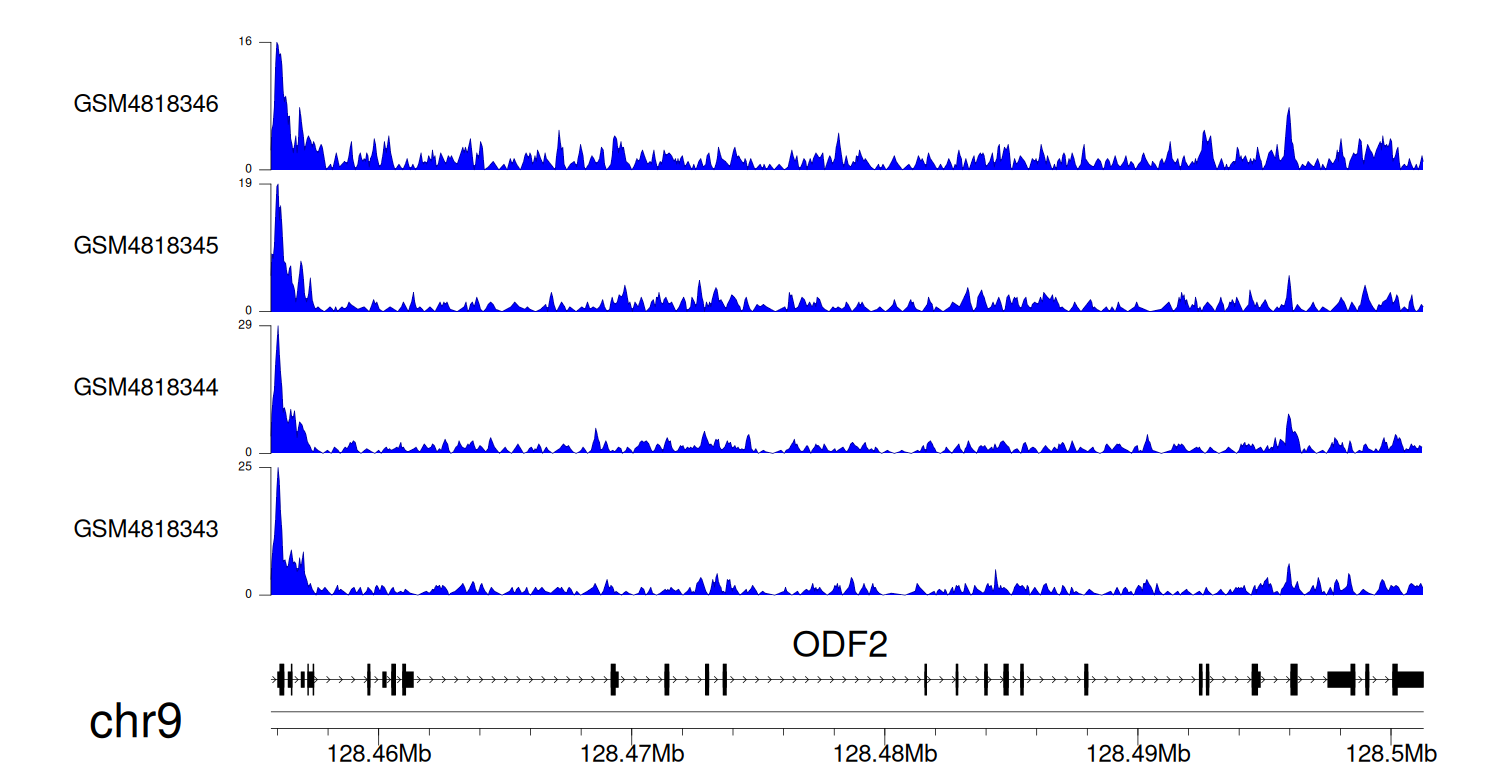
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information