Gene Information
|
Gene Name
|
PARN |
|
Gene ID
|
5073
|
|
Gene Full Name
|
poly(A)-specific ribonuclease |
|
Gene Alias
|
DAN|DKCB6|PFBMFT4 |
|
Transcripts
|
ENSG00000140694
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nuclear speckles; 
|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Potential drug targets, Predicted intracellular proteins |
|
Uniport_ID
|
O95453
|
|
HGNC ID
|
HGNC:8609
|
|
OMIM ID
|
604212 |
|
Summary
|
The protein encoded by this gene is a 3'-exoribonuclease, with similarity to the RNase D family of 3'-exonucleases. It prefers poly(A) as the substrate, hence, efficiently degrades poly(A) tails of mRNAs. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs. This protein is also involved in silencing of certain maternal mRNAs during oocyte maturation and early embryonic development, as well as in nonsense-mediated decay (NMD) of mRNAs that contain premature stop codons. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2008] |
Target gene [PARN] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1041147 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041148 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041149 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041150 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041151 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041152 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041153 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041154 |
chr16 |
1 |
2 |
0 |
3 |
View |
| 1041155 |
chr16 |
2 |
1 |
0 |
3 |
View |
| 1041156 |
chr16 |
2 |
1 |
0 |
3 |
View |
Target gene [PARN] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - PARN peak across samples
|
Peak Plot
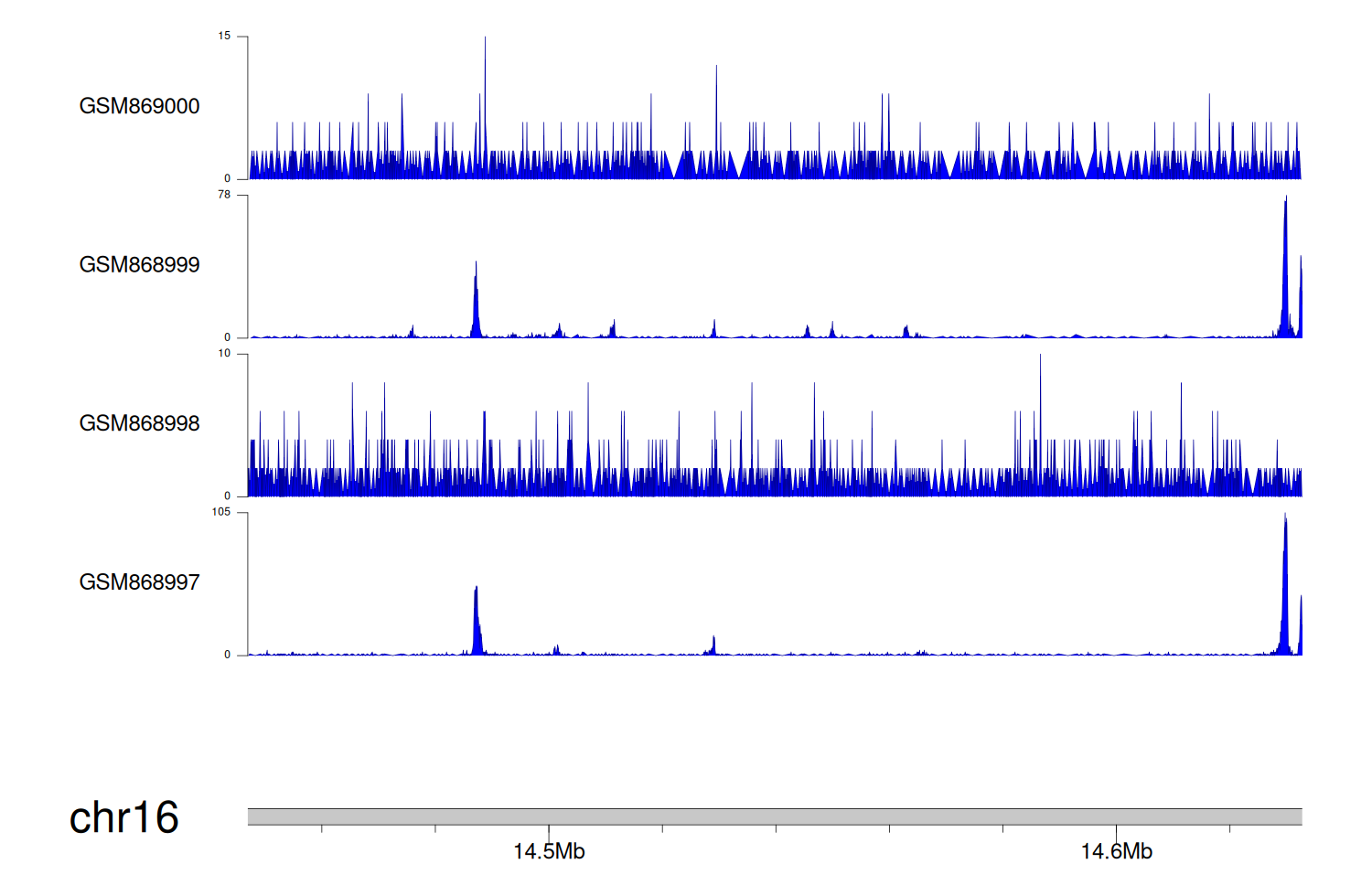
|
> Dataset: GSE68402 - PARN peak across samples
|
Peak Plot
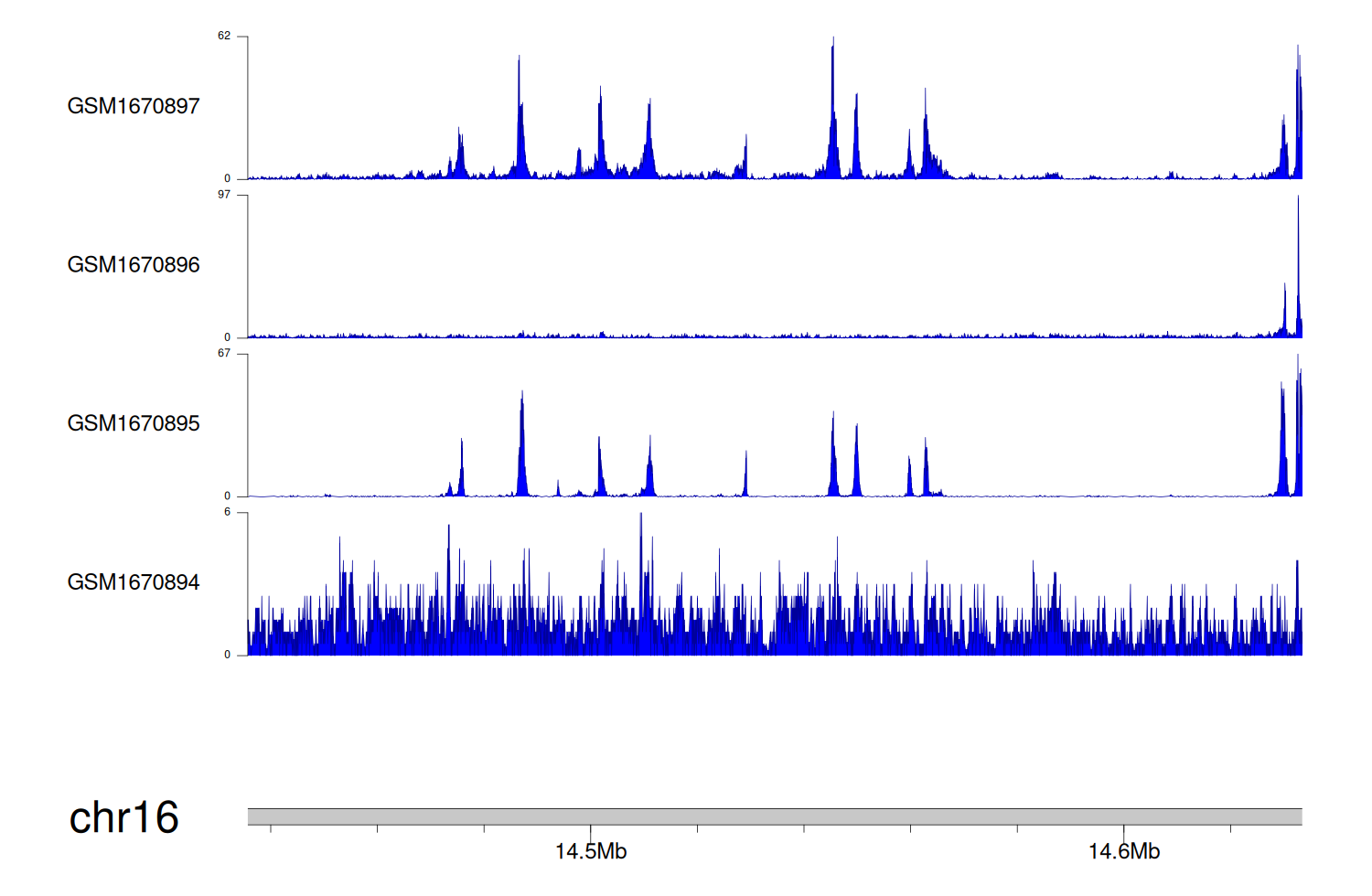
|
> Dataset: GSE270130 - PARN peak across samples
|
Peak Plot
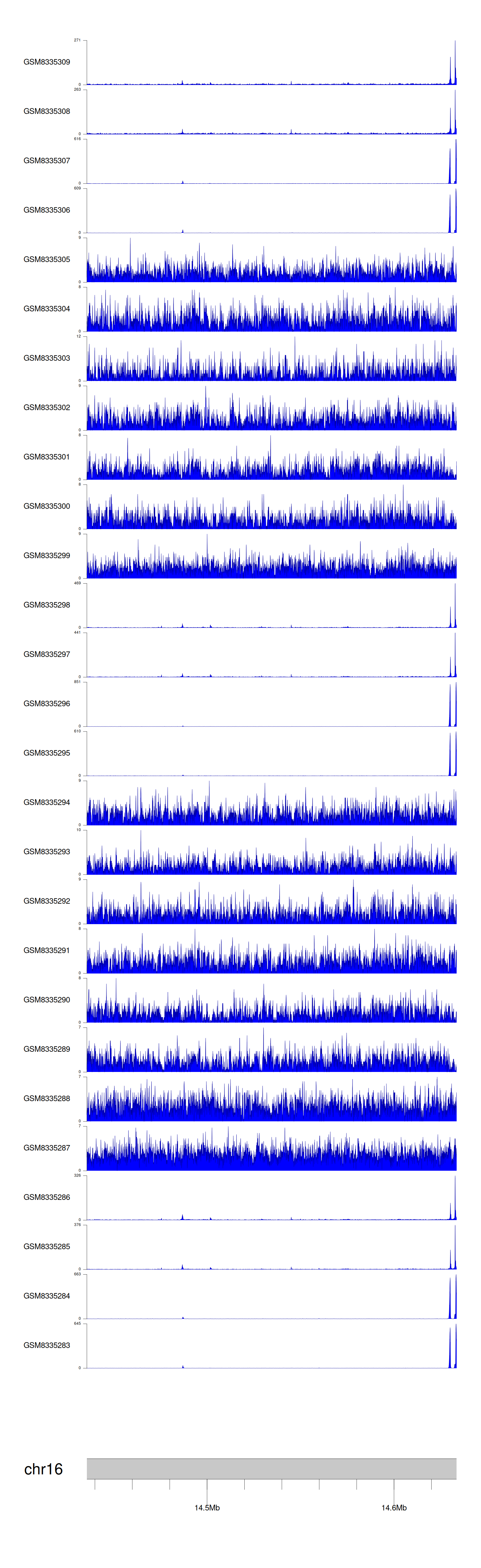
|
> Dataset: GSE100400 - PARN peak across samples
|
Peak Plot

|
> Dataset: GSE131257 - PARN peak across samples
|
Peak Plot
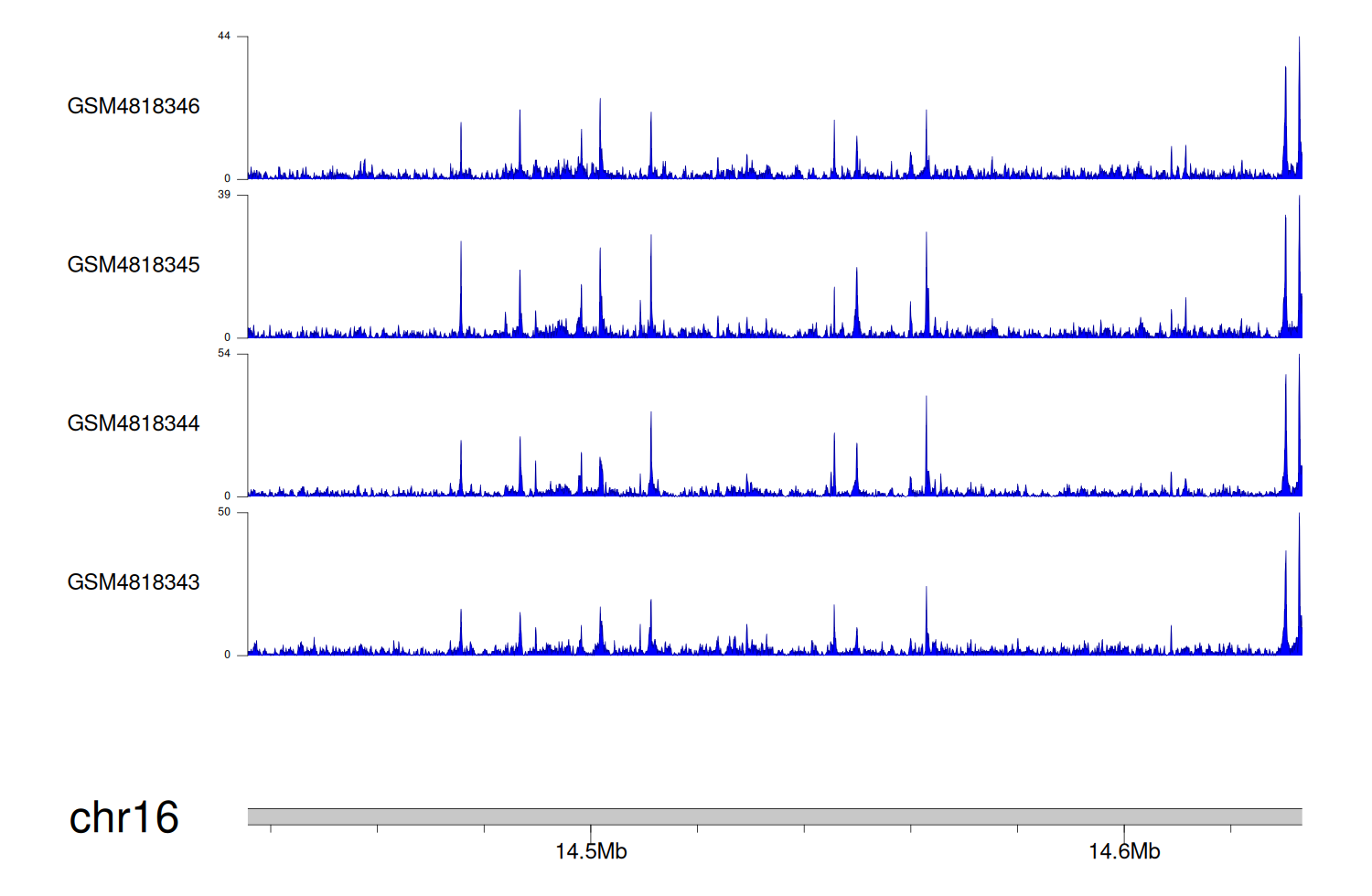
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information