Gene Information
|
Gene Name
|
PIKFYVE |
|
Gene ID
|
200576
|
|
Gene Full Name
|
phosphoinositide kinase, FYVE-type zinc finger containing |
|
Gene Alias
|
CFD|FAB1|HEL37|PIP5K|PIP5K3|ZFYVE29 |
|
Transcripts
|
ENSG00000115020
|
|
Virus
|
HTLV1 |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Potential drug targets, Predicted intracellular proteins |
|
Uniport_ID
|
Q9Y2I7
|
|
HGNC ID
|
HGNC:23785
|
|
OMIM ID
|
609414 |
|
Summary
|
Phosphorylated derivatives of phosphatidylinositol (PtdIns) regulate cytoskeletal functions, membrane trafficking, and receptor signaling by recruiting protein complexes to cell- and endosomal-membranes. Humans have multiple PtdIns proteins that differ by the degree and position of phosphorylation of the inositol ring. This gene encodes an enzyme (PIKfyve; also known as phosphatidylinositol-3-phosphate 5-kinase type III or PIPKIII) that phosphorylates the D-5 position in PtdIns and phosphatidylinositol-3-phosphate (PtdIns3P) to make PtdIns5P and PtdIns(3,5)biphosphate. The D-5 position also can be phosphorylated by type I PtdIns4P-5-kinases (PIP5Ks) that are encoded by distinct genes and preferentially phosphorylate D-4 phosphorylated PtdIns. In contrast, PIKfyve preferentially phosphorylates D-3 phosphorylated PtdIns. In addition to being a lipid kinase, PIKfyve also has protein kinase activity. PIKfyve regulates endomembrane homeostasis and plays a role in the biogenesis of endosome carrier vesicles from early endosomes. The protein plays a key role in cell entry of ebola virus and SARS-CoV-2 by endocytosis Mutations in this gene cause corneal fleck dystrophy (CFD); an autosomal dominant disorder characterized by numerous small white flecks present in all layers of the corneal stroma. Histologically, these flecks appear to be keratocytes distended with lipid and mucopolysaccharide filled intracytoplasmic vacuoles. [provided by RefSeq, Jul 2021] |
Target gene [PIKFYVE] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 6002781 |
chr2 |
8 |
202 |
0 |
210 |
View |
Target gene [PIKFYVE] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE189602
|
Methylation profiling (Array) |
4 |
Infinium MethylationEPIC |
|
C GSE94732
|
Chip-seq |
24 |
Illumina NextSeq 500 (Homo sapiens);illumina Genome Analyzer IIx (Homo sapiens) |
|
GSE52244
|
Expression array |
15 |
[HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [probe set (exon) version] |
|
GSE10789
|
Expression array |
6 |
NCI/ATC Hs-OperonV3 |
|
GSE224047
|
RNA-seq |
10 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE168557
|
Expression array |
6 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Feature Number version) |
|
GSE136189
|
Methylation profiling (Array) |
40 |
Illumina HumanMethylation450 BeadChip (HumanMethylation450_15017482);Illumina Infinium HumanMethylation850 BeadChip |
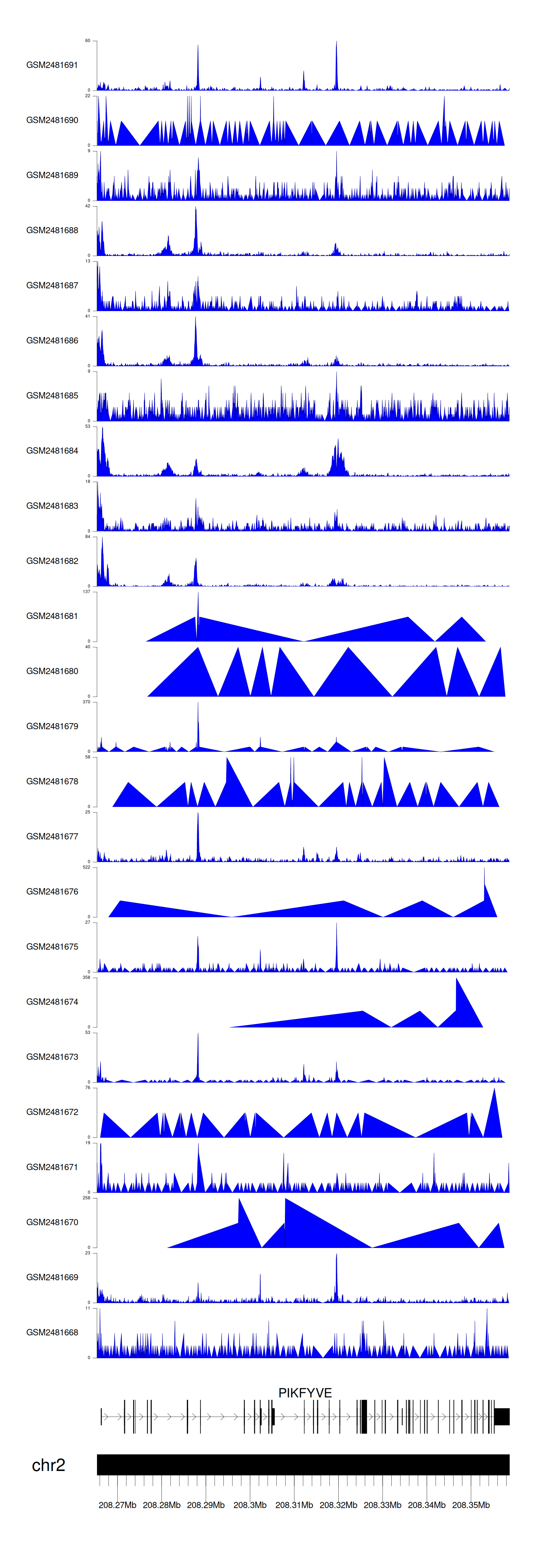
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE94732 - PIKFYVE peak across samples
|
Peak Plot
|
|
|
 HTLV1 Target gene Detail Information
HTLV1 Target gene Detail Information