Gene Information
|
Gene Name
|
PRKG1 |
|
Gene ID
|
5592
|
|
Gene Full Name
|
protein kinase cGMP-dependent 1 |
|
Gene Alias
|
AAT8|PKG|PKG1|PRKG1B|PRKGR1B|cGK|cGK 1|cGK1|cGKI|cGKI-BETA|cGKI-alpha |
|
Transcripts
|
ENSG00000185532
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Vesicles, Cytosol; 
|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Plasma proteins, Potential drug targets, Predicted intracellular proteins |
|
Uniport_ID
|
Q13976
|
|
HGNC ID
|
HGNC:9414
|
|
OMIM ID
|
176894 |
|
Summary
|
Mammals have three different isoforms of cyclic GMP-dependent protein kinase (Ialpha, Ibeta, and II). These PRKG isoforms act as key mediators of the nitric oxide/cGMP signaling pathway and are important components of many signal transduction processes in diverse cell types. This PRKG1 gene on human chromosome 10 encodes the soluble Ialpha and Ibeta isoforms of PRKG by alternative transcript splicing. A separate gene on human chromosome 4, PRKG2, encodes the membrane-bound PRKG isoform II. The PRKG1 proteins play a central role in regulating cardiovascular and neuronal functions in addition to relaxing smooth muscle tone, preventing platelet aggregation, and modulating cell growth. This gene is most strongly expressed in all types of smooth muscle, platelets, cerebellar Purkinje cells, hippocampal neurons, and the lateral amygdala. Isoforms Ialpha and Ibeta have identical cGMP-binding and catalytic domains but differ in their leucine/isoleucine zipper and autoinhibitory sequences and therefore differ in their dimerization substrates and kinase enzyme activity. [provided by RefSeq, Sep 2011] |
Target gene [PRKG1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1003865 |
chr10 |
48 |
25 |
31 |
104 |
View |
| 1012625 |
chr10 |
0 |
2 |
4 |
6 |
View |
| 1016693 |
chr10 |
1 |
0 |
0 |
1 |
View |
| 1016745 |
chr10 |
1 |
0 |
0 |
1 |
View |
| 1017401 |
chr10 |
0 |
1 |
0 |
1 |
View |
| 1018121 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1019014 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1019015 |
chr10 |
0 |
0 |
0 |
0 |
View |
| 1020010 |
chr10 |
11 |
1 |
0 |
12 |
View |
| 1041815 |
chr10 |
36 |
15 |
111 |
162 |
View |
Target gene [PRKG1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE64877
|
Chip-seq |
2 |
Illumina Genome Analyzer (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - PRKG1 expression across samples
|
Volcano Plot
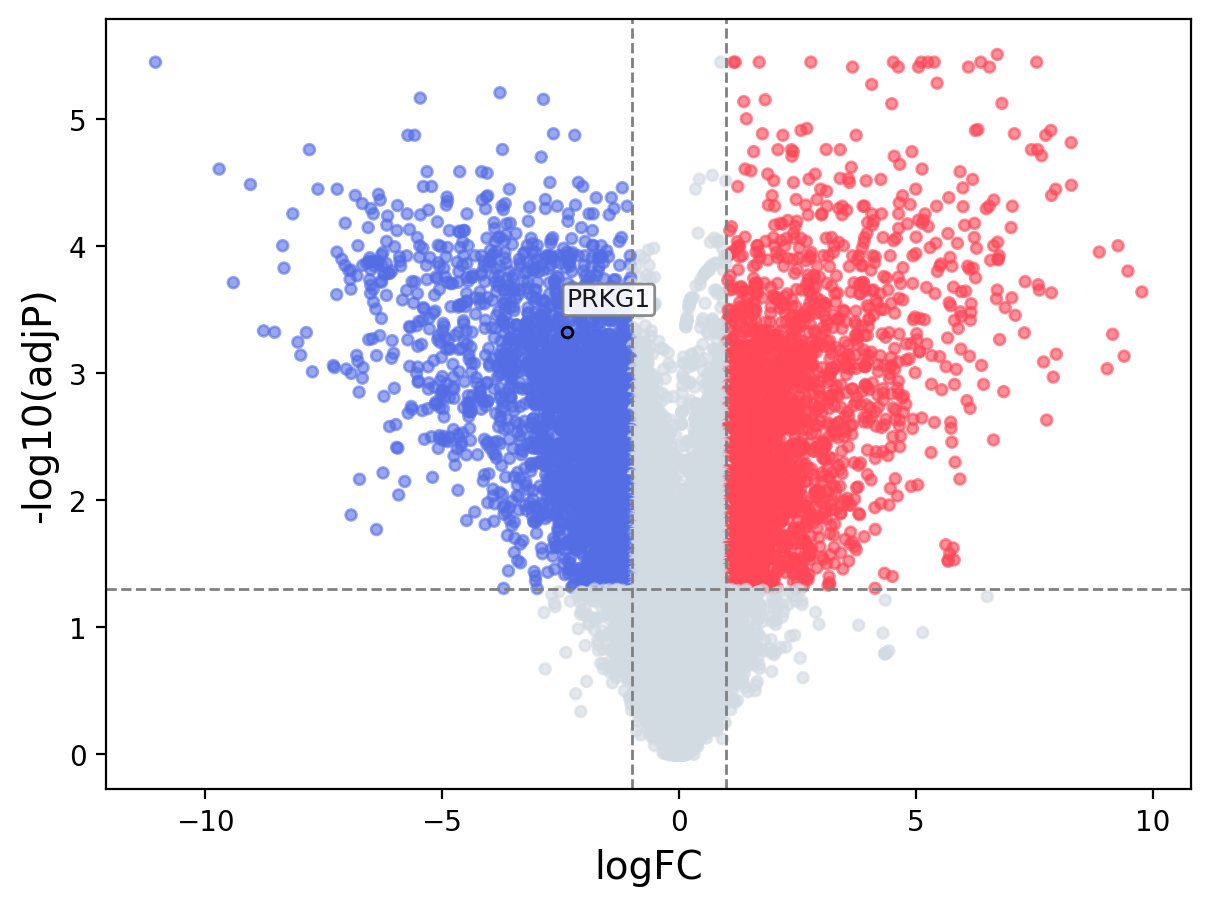
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information