Gene Information
|
Gene Name
|
PTPRB |
|
Gene ID
|
5787
|
|
Gene Full Name
|
protein tyrosine phosphatase receptor type B |
|
Gene Alias
|
HPTP-BETA|HPTPB|PTPB|R-PTP-BETA|VEPTP |
|
Transcripts
|
ENSG00000127329
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Vesicles, Cell Junctions; 
|
|
Membrane Info
|
Cancer-related genes, Enzymes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
P23467
|
|
HGNC ID
|
HGNC:9665
|
|
OMIM ID
|
176882 |
|
Summary
|
The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP contains an extracellular domain, a single transmembrane segment and one intracytoplasmic catalytic domain, thus belongs to receptor type PTP. The extracellular region of this PTP is composed of multiple fibronectin type_III repeats, which was shown to interact with neuronal receptor and cell adhesion molecules, such as contactin and tenascin C. This protein was also found to interact with sodium channels, and thus may regulate sodium channels by altering tyrosine phosphorylation status. The functions of the interaction partners of this protein implicate the roles of this PTP in cell adhesion, neurite growth, and neuronal differentiation. Alternate transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, May 2011] |
Target gene [PTPRB] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1041070 |
chr12 |
2 |
0 |
0 |
2 |
View |
| 1041071 |
chr12 |
1 |
0 |
0 |
1 |
View |
| 1041072 |
chr12 |
1 |
0 |
0 |
1 |
View |
| 1041073 |
chr12 |
1 |
0 |
0 |
1 |
View |
Target gene [PTPRB] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
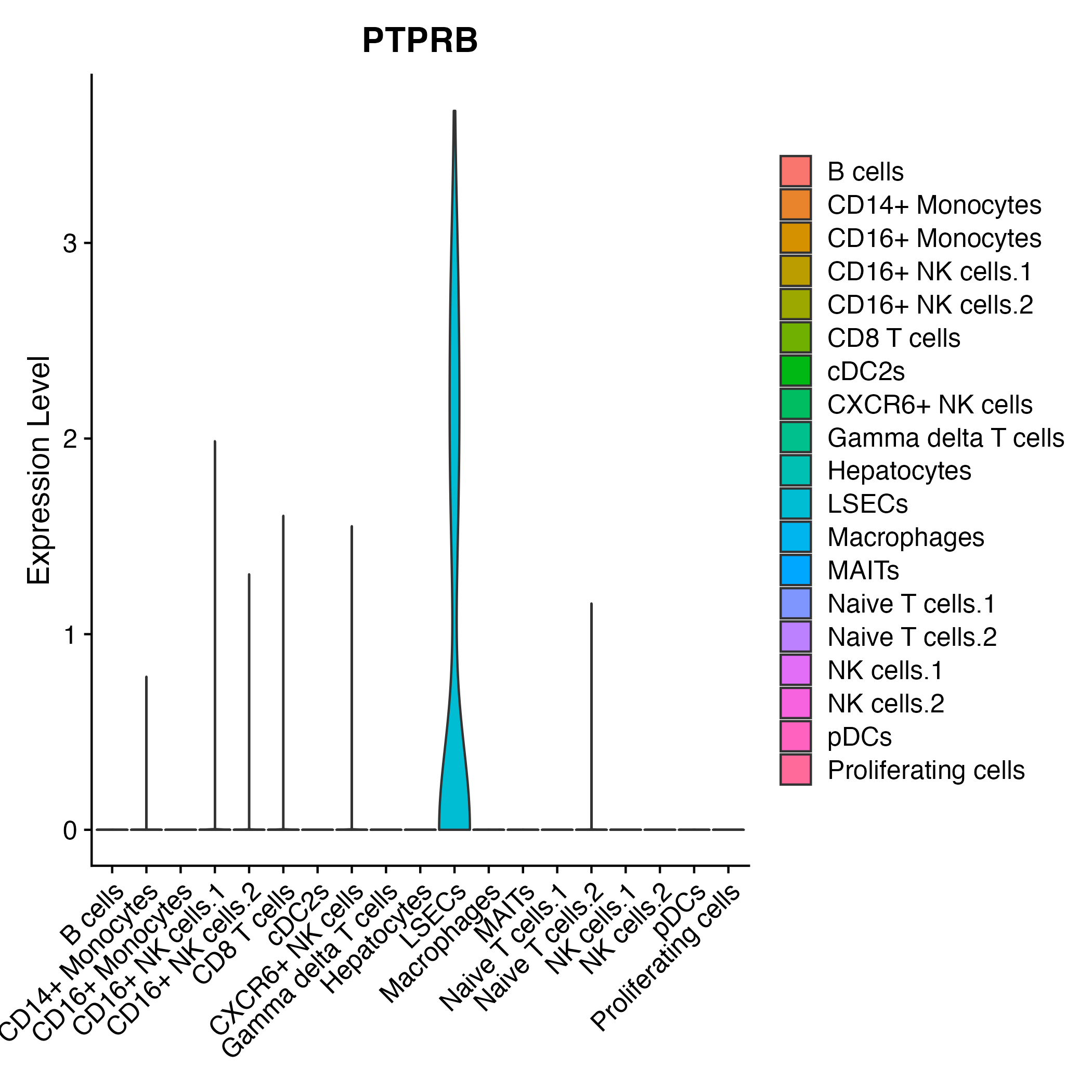
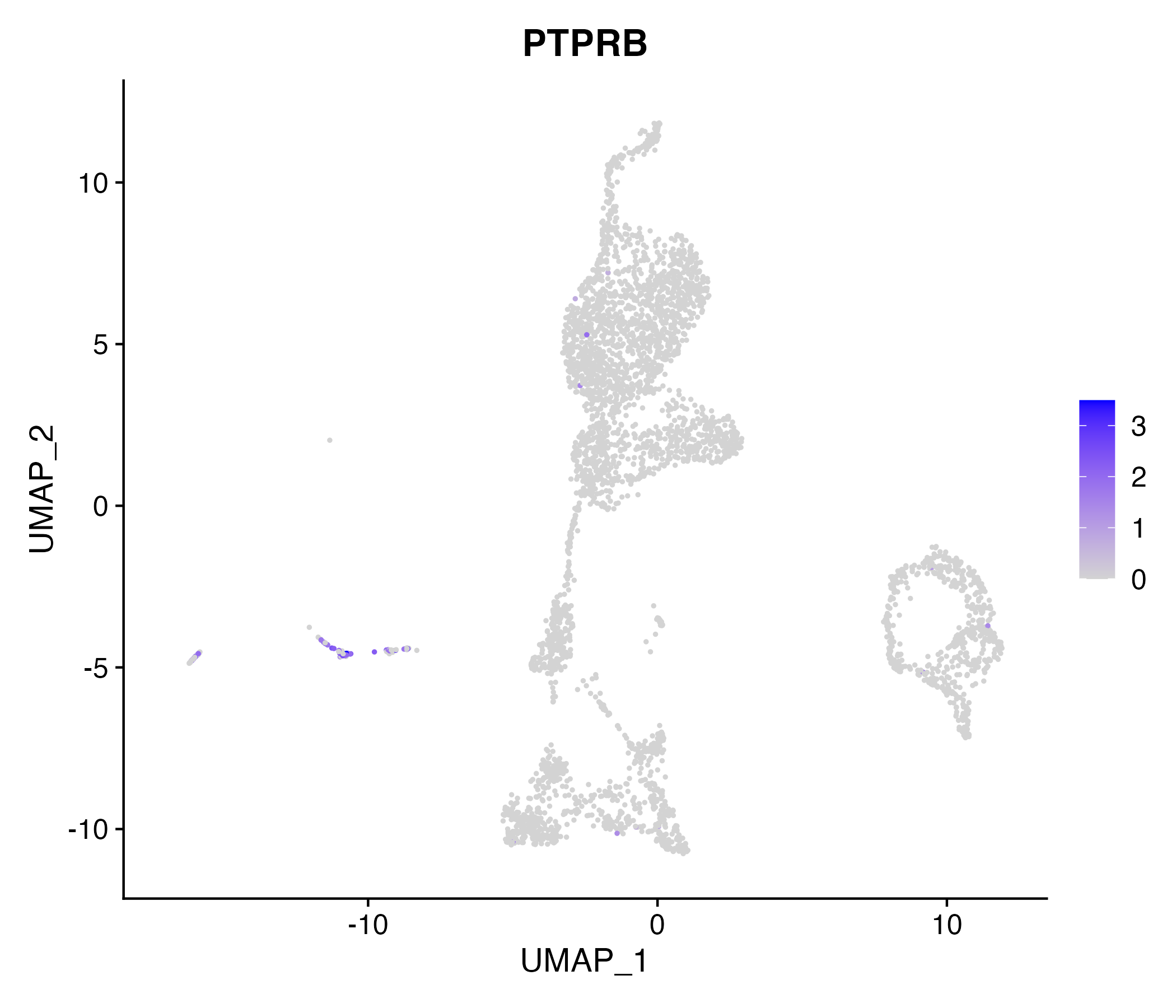
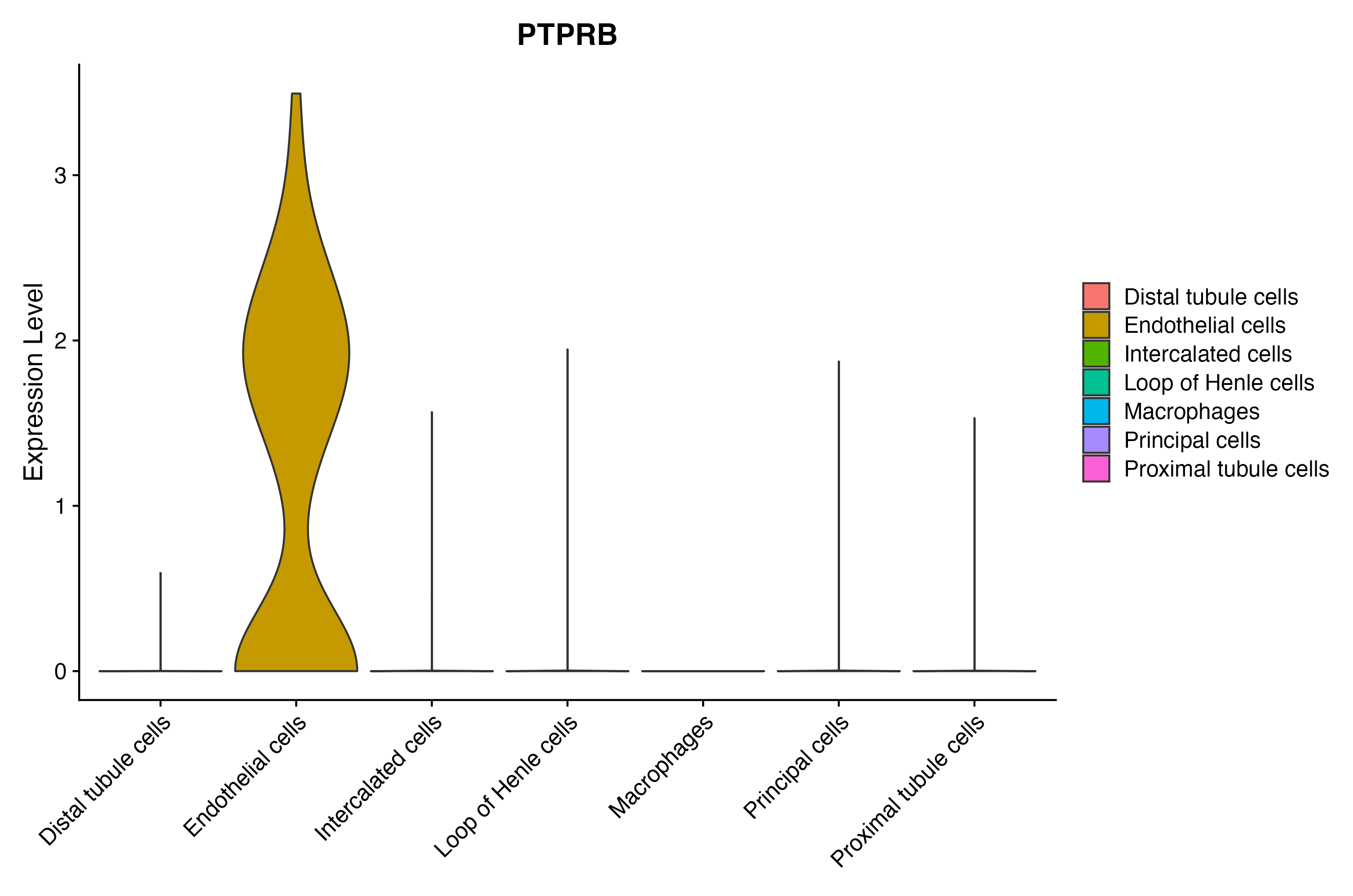
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE252863 - Gene expression in cell subsets
|
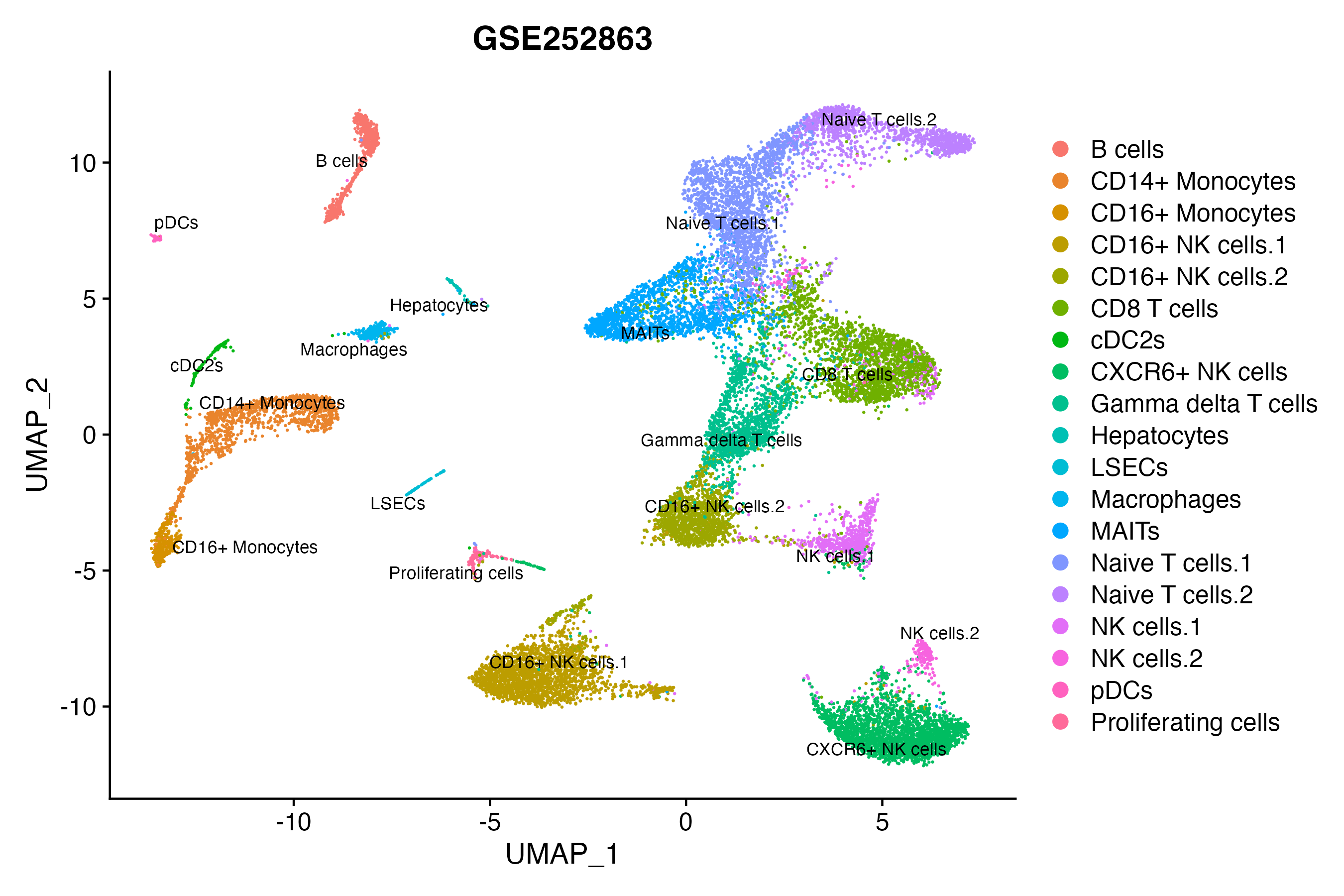
|
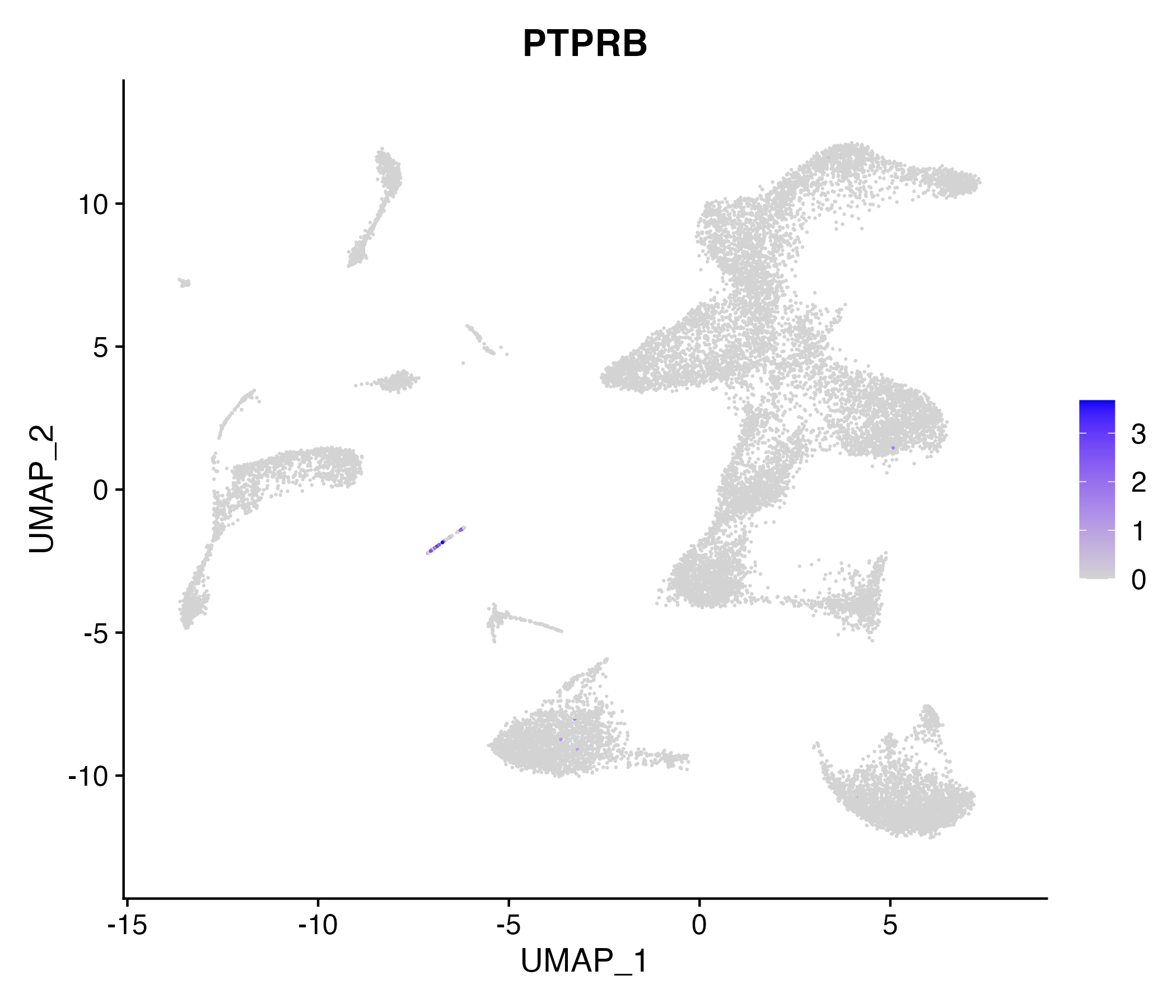
|
|
|
|
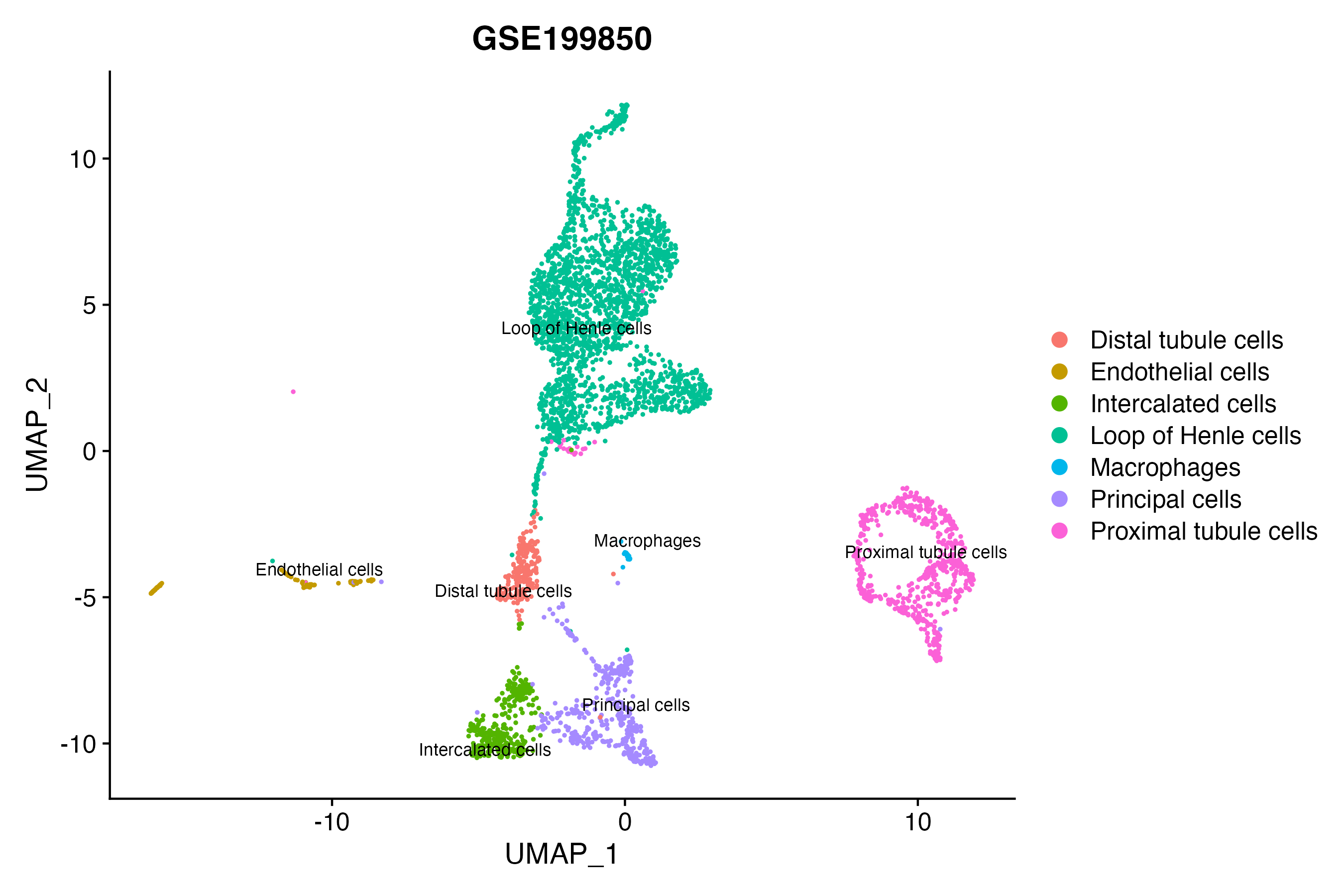
> Dataset: GSE199850 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information