Gene Information
|
Gene Name
|
CAMK2D |
|
Gene ID
|
817
|
|
Gene Full Name
|
calcium/calmodulin dependent protein kinase II delta |
|
Gene Alias
|
CAMKD |
|
Transcripts
|
ENSG00000145349
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane, Primary cilium tip, Primary cilium transition zone, Principal piece;Cell Junctions, Perinuclear theca, Calyx, Connecting piece, End piece; 
|
|
Membrane Info
|
Enzymes, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
Q13557
|
|
HGNC ID
|
HGNC:1462
|
|
OMIM ID
|
607708 |
|
Summary
|
The product of this gene belongs to the serine/threonine protein kinase family and to the Ca(2+)/calmodulin-dependent protein kinase subfamily. Calcium signaling is crucial for several aspects of plasticity at glutamatergic synapses. In mammalian cells, the enzyme is composed of four different chains: alpha, beta, gamma, and delta. The product of this gene is a delta chain. Alternative splicing results in multiple transcript variants encoding distinct isoforms. Distinct isoforms of this chain have different expression patterns.[provided by RefSeq, Nov 2008] |
Target gene [CAMK2D] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1009740 |
chr4 |
12 |
2 |
0 |
14 |
View |
| 1020429 |
chr4 |
4 |
12 |
55 |
71 |
View |
| 1022499 |
chr4 |
12 |
2 |
0 |
14 |
View |
| 1024233 |
chr4 |
0 |
0 |
0 |
0 |
View |
| 1024354 |
chr4 |
53 |
0 |
0 |
53 |
View |
| 1024506 |
chr4 |
3 |
0 |
0 |
3 |
View |
| 1042868 |
chr4 |
16 |
2 |
0 |
18 |
View |
Target gene [CAMK2D] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE64877
|
Chip-seq |
2 |
Illumina Genome Analyzer (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
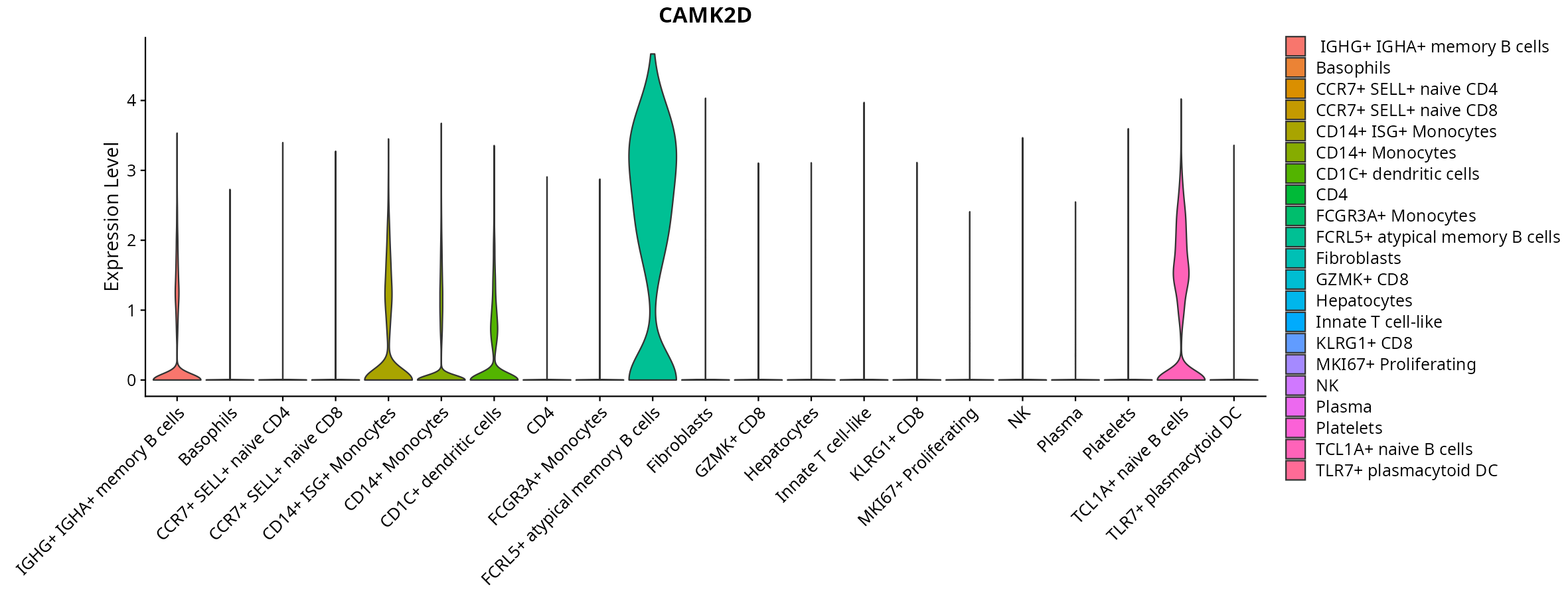
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE247322 - Gene expression in cell subsets
|
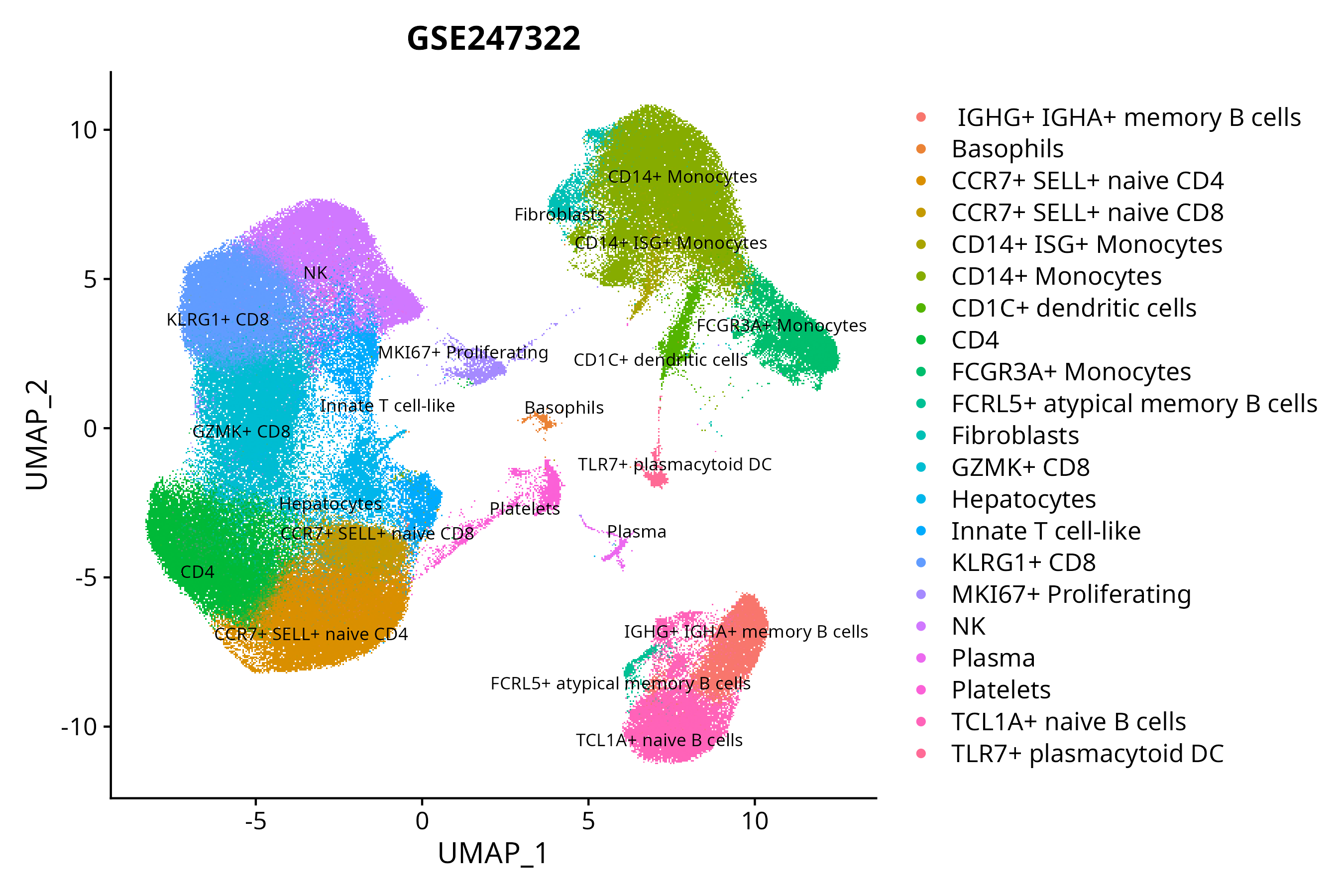
|
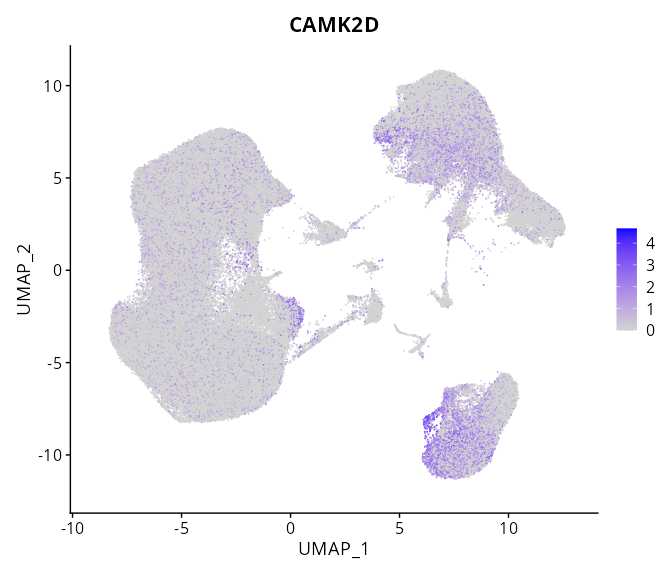
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information