Gene Information
|
Gene Name
|
FHIT |
|
Gene ID
|
2272
|
|
Gene Full Name
|
fragile histidine triad diadenosine triphosphatase |
|
Gene Alias
|
AP3Aase|FRA3B |
|
Transcripts
|
ENSG00000189283
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoli fibrillar center, Plasma membrane; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Potential drug targets, Predicted intracellular proteins |
|
Uniport_ID
|
P49789
|
|
HGNC ID
|
HGNC:3701
|
|
OMIM ID
|
601153 |
|
Summary
|
The protein encoded by this gene is a P1-P3-bis(5'-adenosyl) triphosphate hydrolase involved in purine metabolism. This gene encompasses the common fragile site FRA3B on chromosome 3, where carcinogen-induced damage can lead to translocations and aberrant transcripts. In fact, aberrant transcripts from this gene have been found in about half of all esophageal, stomach, and colon carcinomas. The encoded protein is also a tumor suppressor, as loss of its activity results in replication stress and DNA damage. [provided by RefSeq, Aug 2017] |
Target gene [FHIT] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1004550 |
chr3 |
67 |
6 |
4 |
77 |
View |
| 1005083 |
chr3 |
24 |
27 |
10 |
61 |
View |
| 1006191 |
chr3 |
4 |
28 |
10 |
42 |
View |
| 1014029 |
chr3 |
3 |
0 |
1 |
4 |
View |
| 1017368 |
chr3 |
1 |
0 |
0 |
1 |
View |
| 1017506 |
chr3 |
0 |
0 |
0 |
0 |
View |
| 1017617 |
chr3 |
0 |
0 |
0 |
0 |
View |
| 1018069 |
chr3 |
0 |
0 |
0 |
0 |
View |
| 1019855 |
chr3 |
0 |
0 |
0 |
0 |
View |
| 1020743 |
chr3 |
0 |
0 |
0 |
0 |
View |
| 1020744 |
chr3 |
67 |
6 |
4 |
77 |
View |
| 1041355 |
chr3 |
1 |
0 |
0 |
1 |
View |
| 1042761 |
chr3 |
3 |
4 |
0 |
7 |
View |
| 1042762 |
chr3 |
0 |
1 |
0 |
1 |
View |
| 1042763 |
chr3 |
139 |
10 |
4 |
153 |
View |
| 1042764 |
chr3 |
0 |
0 |
0 |
0 |
View |
Target gene [FHIT] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
E C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
E GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE100400 - FHIT expression across samples
|
Volcano Plot
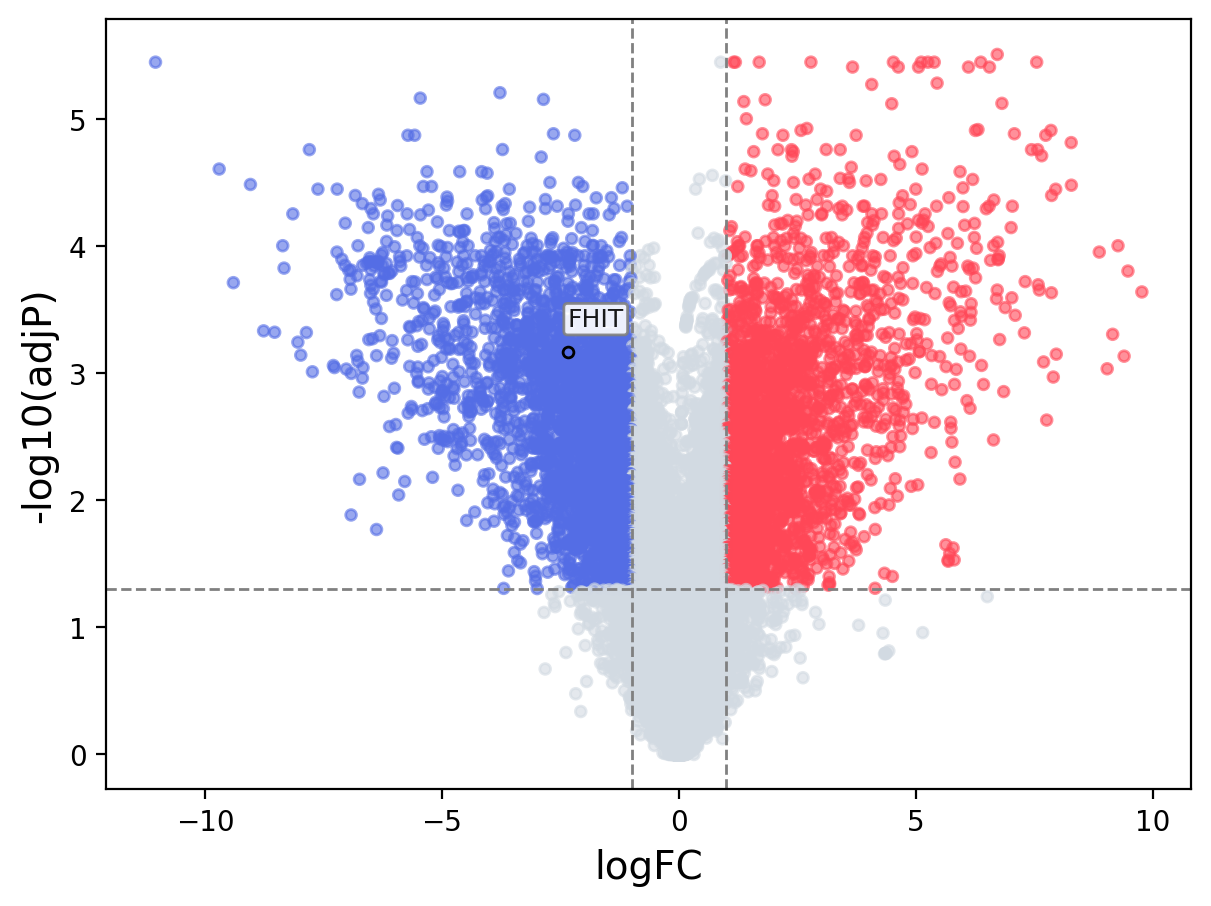
|
Bar Plot
|
> Dataset: GSE94660 - FHIT expression across samples
|
Volcano Plot
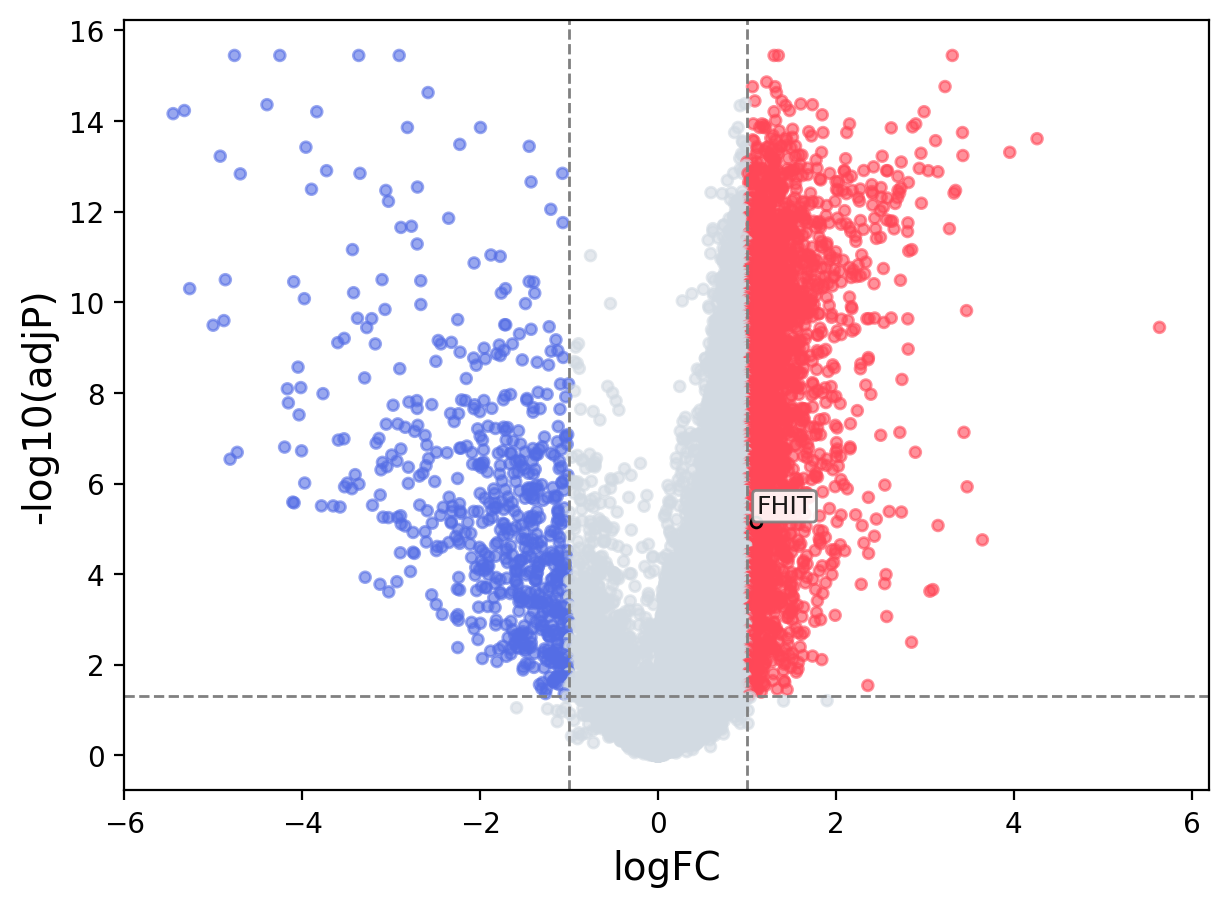
|
Bar Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information