Gene Information
|
Gene Name
|
IKZF1 |
|
Gene ID
|
10320
|
|
Gene Full Name
|
IKAROS family zinc finger 1 |
|
Gene Alias
|
CVID13|Hs.54452|IK1|IKAROS|LYF1|LyF-1|PPP1R92|PRO0758|ZNFN1A1 |
|
Transcripts
|
ENSG00000185811
|
|
Virus
|
HTLV1 |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Predicted intracellular proteins, Predicted membrane proteins, Transcription factors |
|
Uniport_ID
|
Q13422
|
|
HGNC ID
|
HGNC:13176
|
|
OMIM ID
|
603023 |
|
Summary
|
This gene encodes a transcription factor that belongs to the family of zinc-finger DNA-binding proteins associated with chromatin remodeling. The expression of this protein is restricted to the fetal and adult hemo-lymphopoietic system, and it functions as a regulator of lymphocyte differentiation. Several alternatively spliced transcript variants encoding different isoforms have been described for this gene. Most isoforms share a common C-terminal domain, which contains two zinc finger motifs that are required for hetero- or homo-dimerization, and for interactions with other proteins. The isoforms, however, differ in the number of N-terminal zinc finger motifs that bind DNA and in nuclear localization signal presence, resulting in members with and without DNA-binding properties. Only a few isoforms contain the requisite three or more N-terminal zinc motifs that confer high affinity binding to a specific core DNA sequence element in the promoters of target genes. The non-DNA-binding isoforms are largely found in the cytoplasm, and are thought to function as dominant-negative factors. Overexpression of some dominant-negative isoforms have been associated with B-cell malignancies, such as acute lymphoblastic leukemia (ALL). [provided by RefSeq, May 2014] |
Target gene [IKZF1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
Target gene [IKZF1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE189602
|
Methylation profiling (Array) |
4 |
Infinium MethylationEPIC |
|
C GSE94732
|
Chip-seq |
24 |
Illumina NextSeq 500 (Homo sapiens);illumina Genome Analyzer IIx (Homo sapiens) |
|
GSE52244
|
Expression array |
15 |
[HuEx-1_0-st] Affymetrix Human Exon 1.0 ST Array [probe set (exon) version] |
|
GSE19080
|
Expression array |
38 |
Homo sapiens 3K Immunoarray2 HTLV-1 JH |
|
GSE224047
|
RNA-seq |
10 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE168557
|
Expression array |
6 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Feature Number version) |
|
GSE136189
|
Methylation profiling (Array) |
40 |
Illumina HumanMethylation450 BeadChip (HumanMethylation450_15017482);Illumina Infinium HumanMethylation850 BeadChip |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE94732 - IKZF1 peak across samples
|
Peak Plot
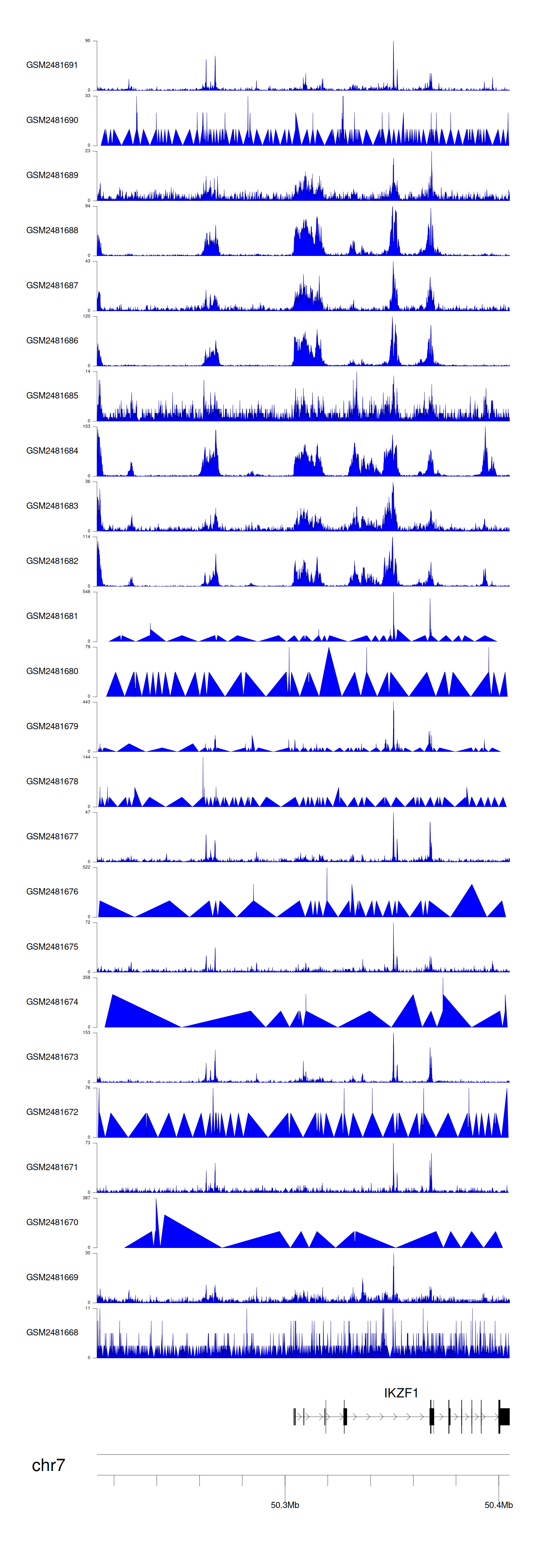
|
|
|
 HTLV1 Target gene Detail Information
HTLV1 Target gene Detail Information