Gene Information
|
Gene Name
|
KMT2A |
|
Gene ID
|
4297
|
|
Gene Full Name
|
lysine methyltransferase 2A |
|
Gene Alias
|
ALL-1|ALL1|CXXC7|GAS7|HRX|HTRX|HTRX1|MLL|MLL1|MLL1A|TRX1|WDSTS |
|
Transcripts
|
ENSG00000118058
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Plasma proteins, Potential drug targets, Predicted intracellular proteins, Transcription factors |
|
Uniport_ID
|
Q03164
|
|
HGNC ID
|
HGNC:7132
|
|
OMIM ID
|
159555 |
|
Summary
|
This gene encodes a transcriptional coactivator that plays an essential role in regulating gene expression during early development and hematopoiesis. The encoded protein contains multiple conserved functional domains. One of these domains, the SET domain, is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity which mediates chromatin modifications associated with epigenetic transcriptional activation. This protein is processed by the enzyme Taspase 1 into two fragments, MLL-C and MLL-N. These fragments reassociate and further assemble into different multiprotein complexes that regulate the transcription of specific target genes, including many of the HOX genes. Multiple chromosomal translocations involving this gene are the cause of certain acute lymphoid leukemias and acute myeloid leukemias. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Oct 2010] |
Target gene [KMT2A] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1005216 |
chr11 |
44 |
11 |
3 |
58 |
View |
Target gene [KMT2A] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - KMT2A peak across samples
|
Peak Plot
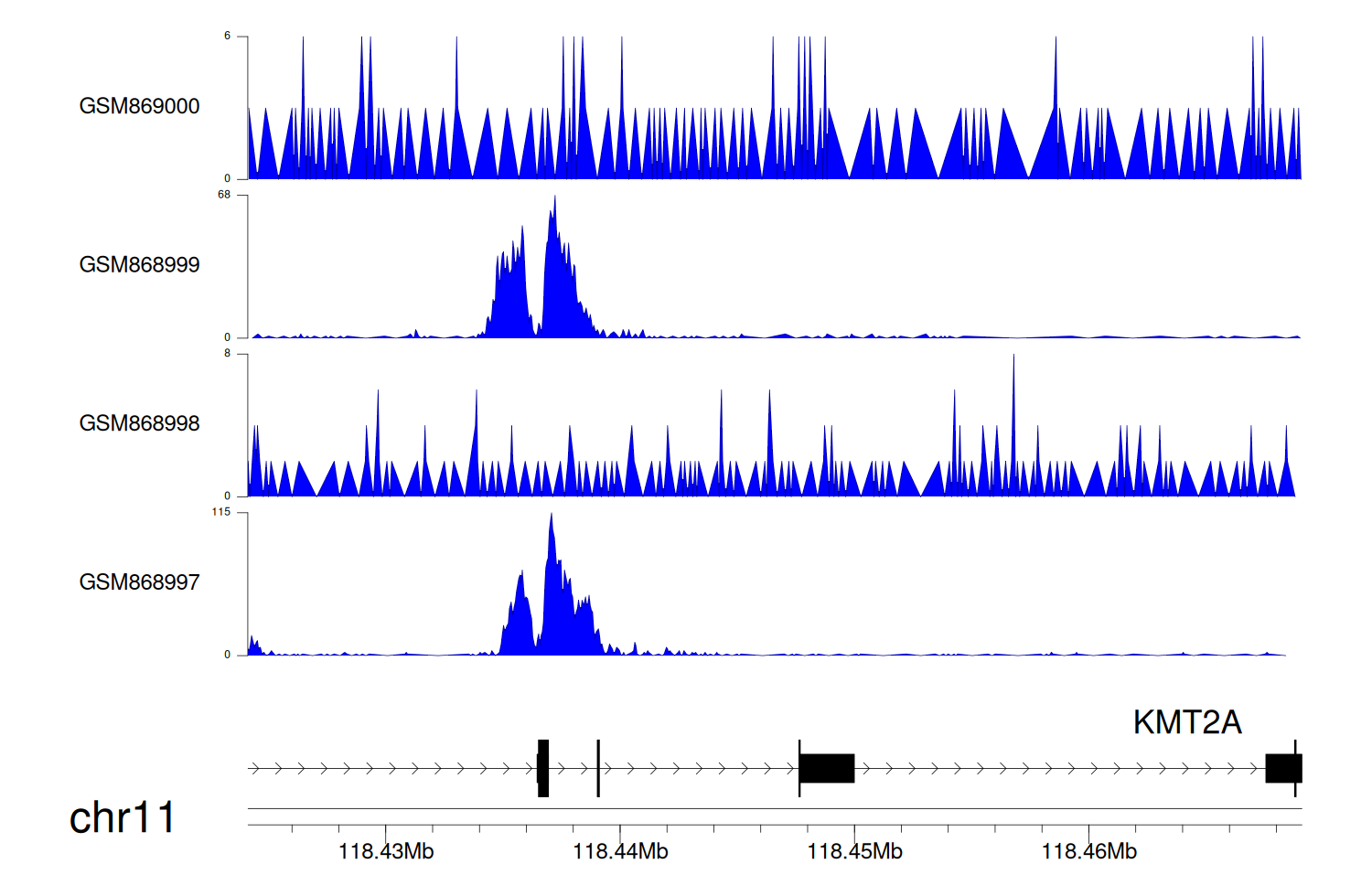
|
> Dataset: GSE68402 - KMT2A peak across samples
|
Peak Plot
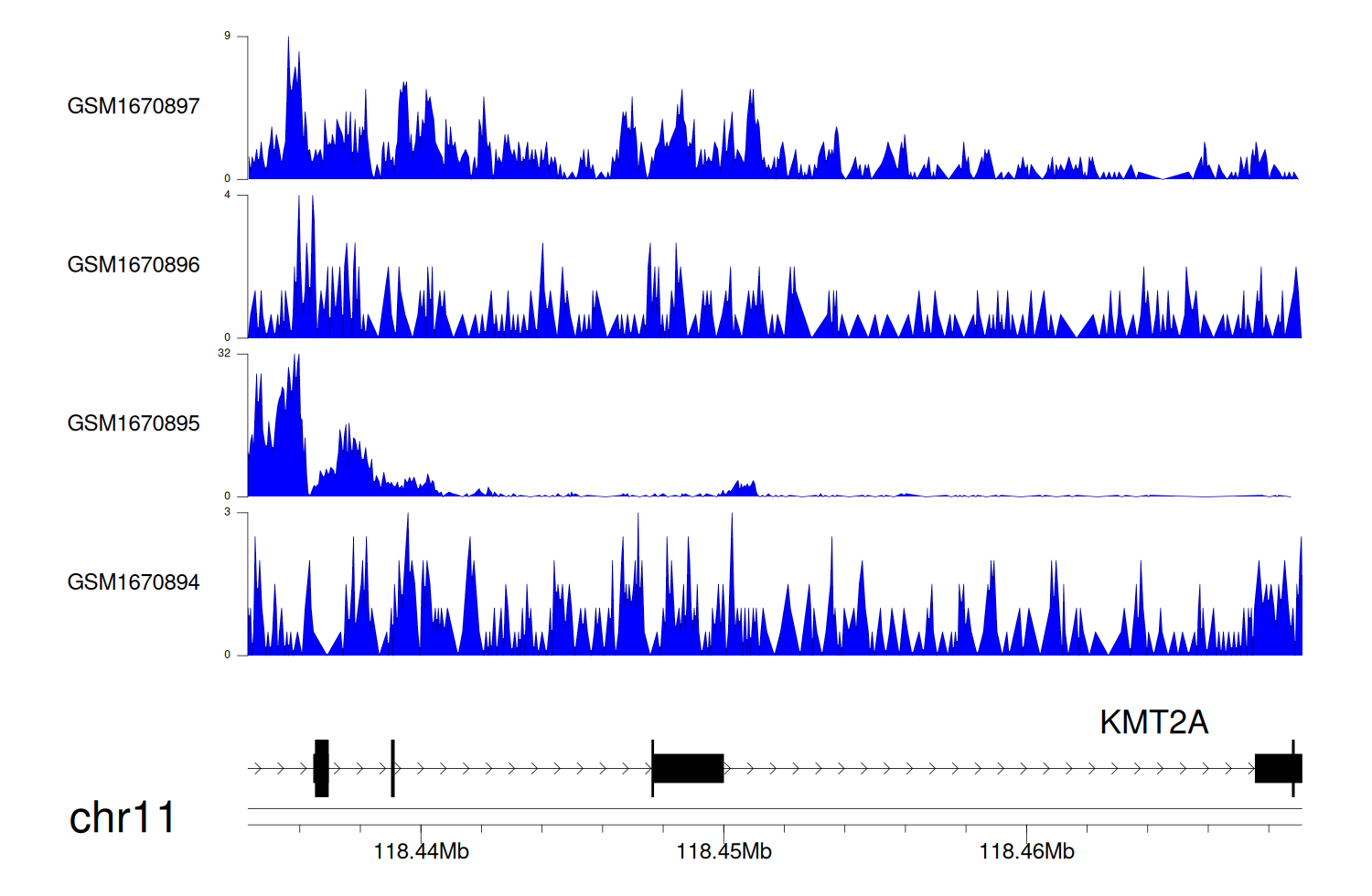
|
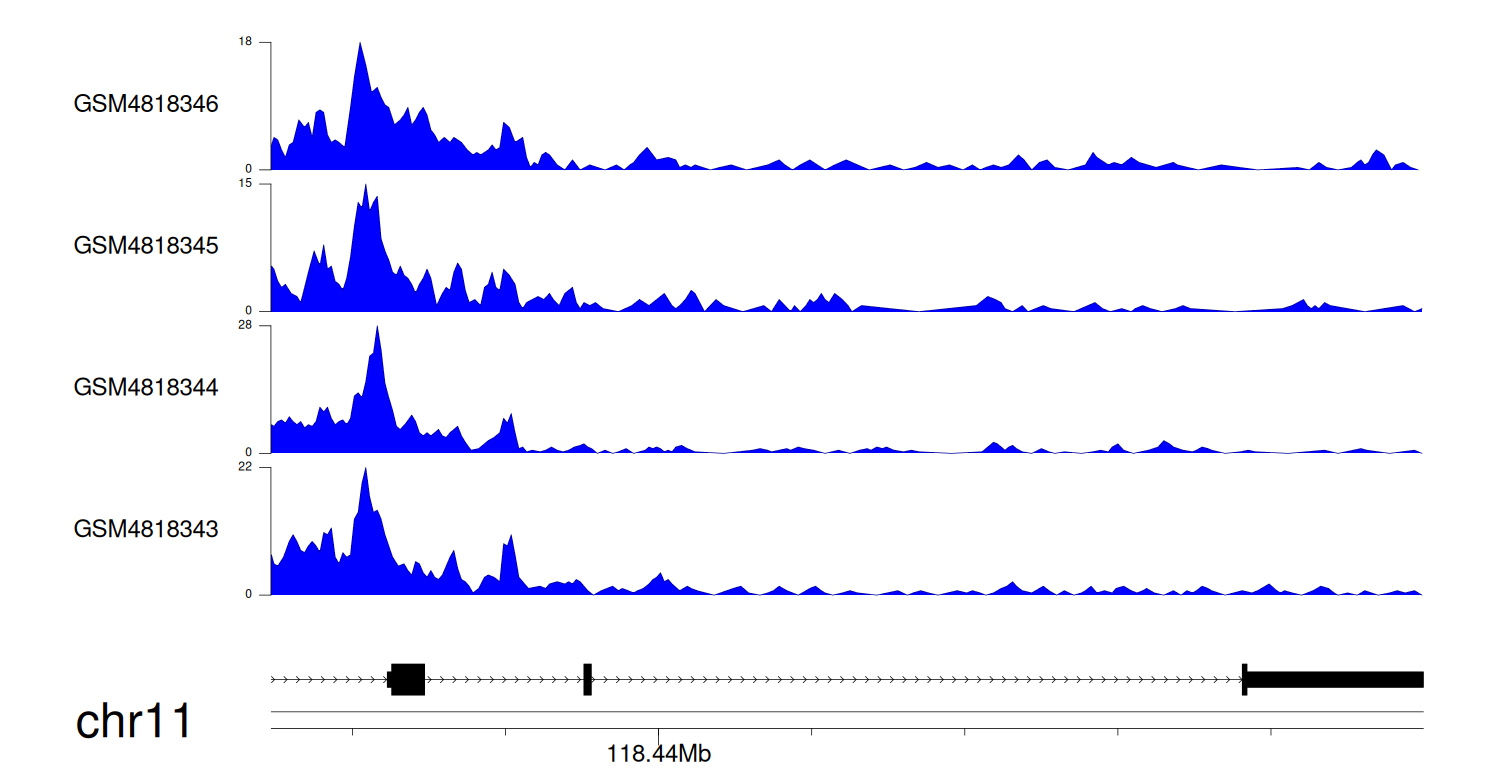
> Dataset: GSE270130 - KMT2A peak across samples
|
Peak Plot
|
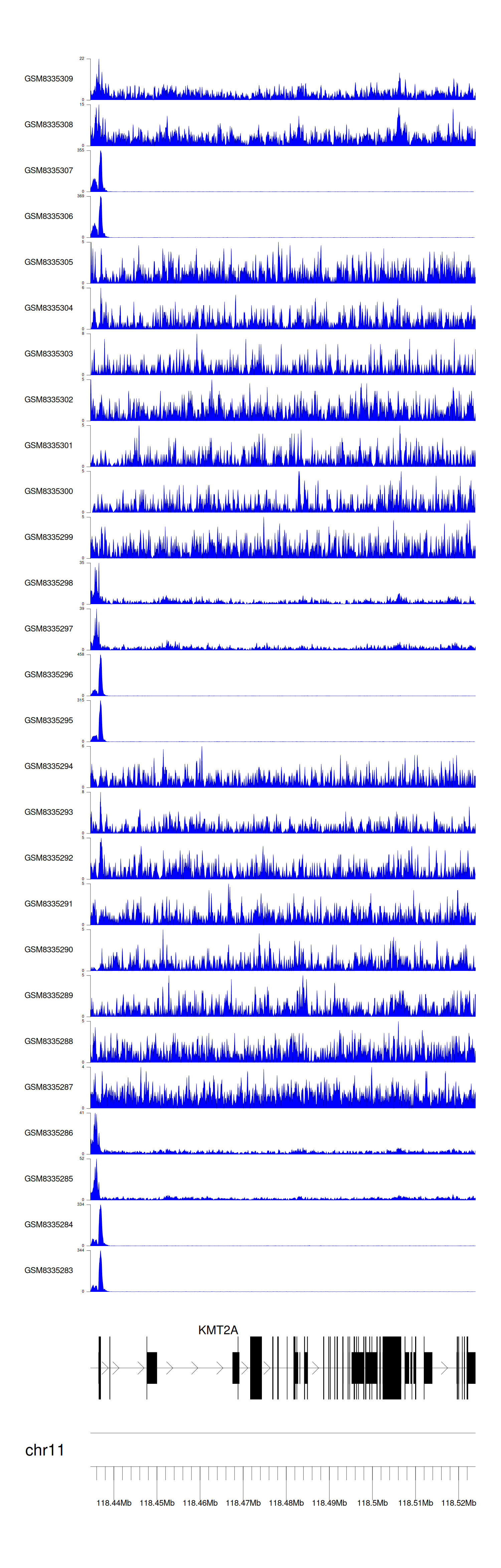
> Dataset: GSE131257 - KMT2A peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information