Gene Information
|
Gene Name
|
LRP1 |
|
Gene ID
|
4035
|
|
Gene Full Name
|
LDL receptor related protein 1 |
|
Gene Alias
|
A2MR|APOER|APR|CD91|DDH3|IGFBP-3R|IGFBP3R|IGFBP3R1|KPA|LRP|LRP1A|TGFBR5 |
|
Transcripts
|
ENSG00000123384
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Nucleoli, Cytosol;Intracellular and membrane; 
|
|
Membrane Info
|
Candidate cardiovascular disease genes, CD markers, Disease related genes, Human disease related genes, Plasma proteins, Potential drug targets, Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
Q07954
|
|
HGNC ID
|
HGNC:6692
|
|
OMIM ID
|
107770 |
|
Summary
|
This gene encodes a member of the low-density lipoprotein receptor family of proteins. The encoded preproprotein is proteolytically processed by furin to generate 515 kDa and 85 kDa subunits that form the mature receptor (PMID: 8546712). This receptor is involved in several cellular processes, including intracellular signaling, lipid homeostasis, and clearance of apoptotic cells. In addition, the encoded protein is necessary for the alpha 2-macroglobulin-mediated clearance of secreted amyloid precursor protein and beta-amyloid, the main component of amyloid plaques found in Alzheimer patients. Expression of this gene decreases with age and has been found to be lower than controls in brain tissue from Alzheimer's disease patients. [provided by RefSeq, Oct 2015] |
Target gene [LRP1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1020164 |
chr12 |
9 |
3 |
0 |
12 |
View |
| 1041069 |
chr12 |
17 |
5 |
0 |
22 |
View |
Target gene [LRP1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
S GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
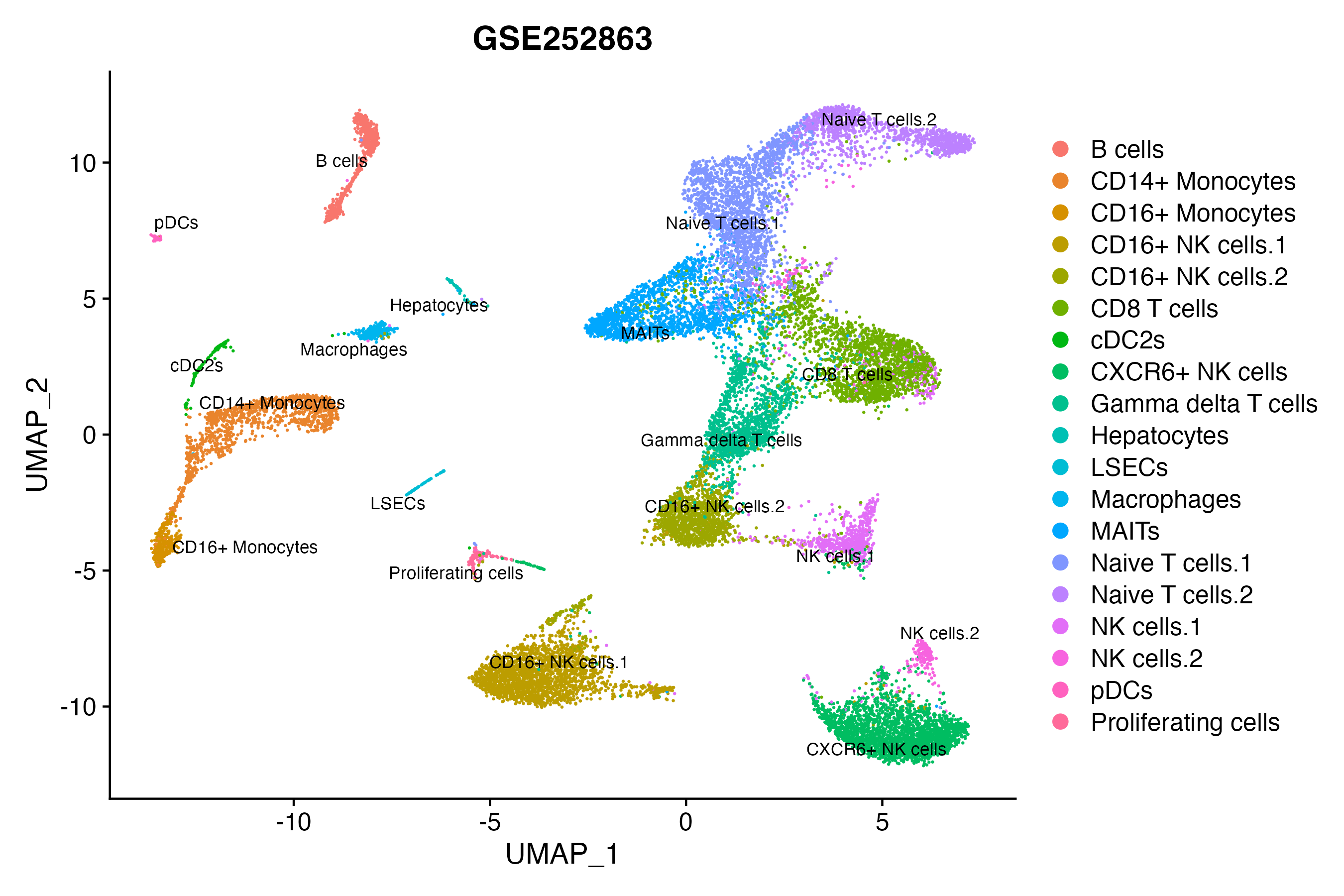
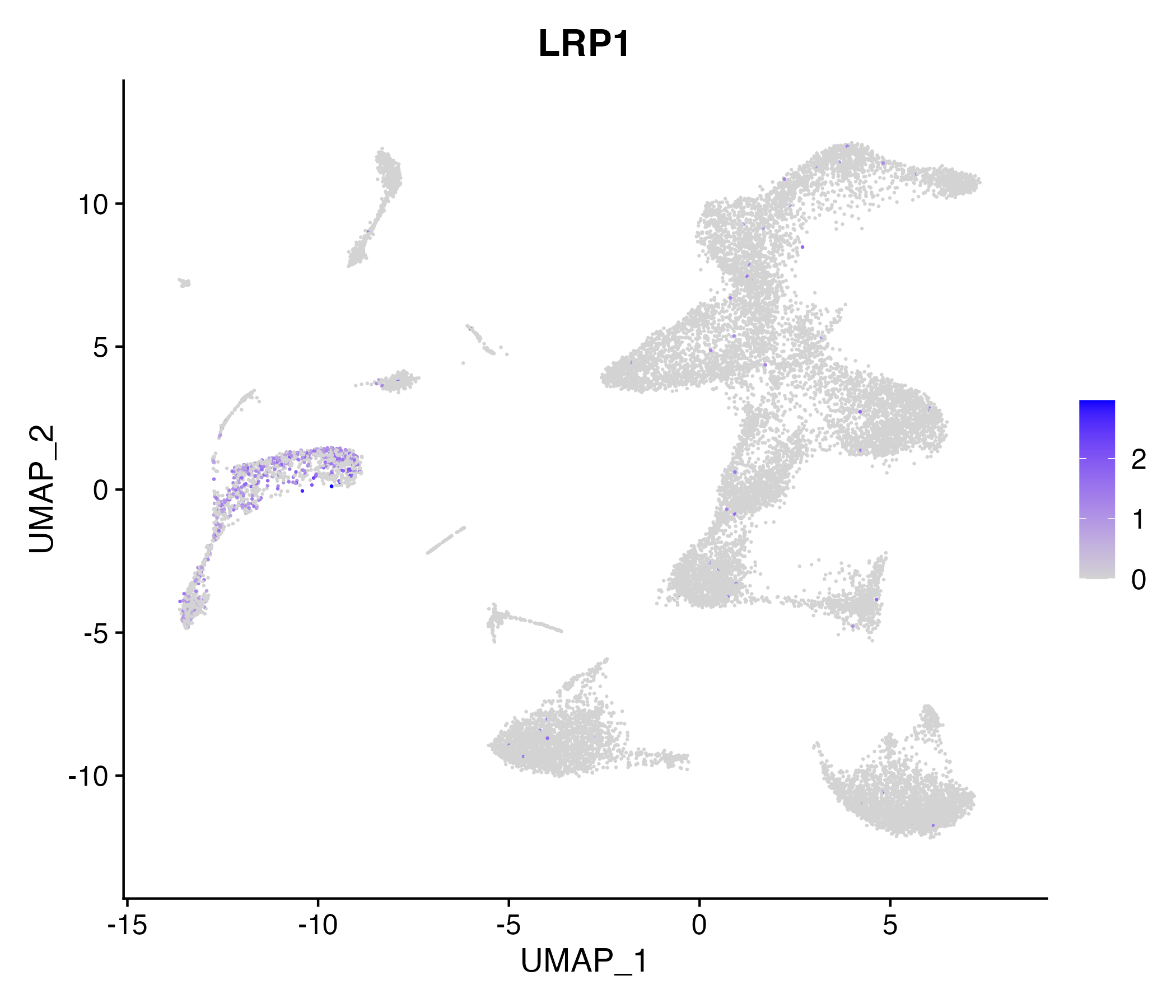
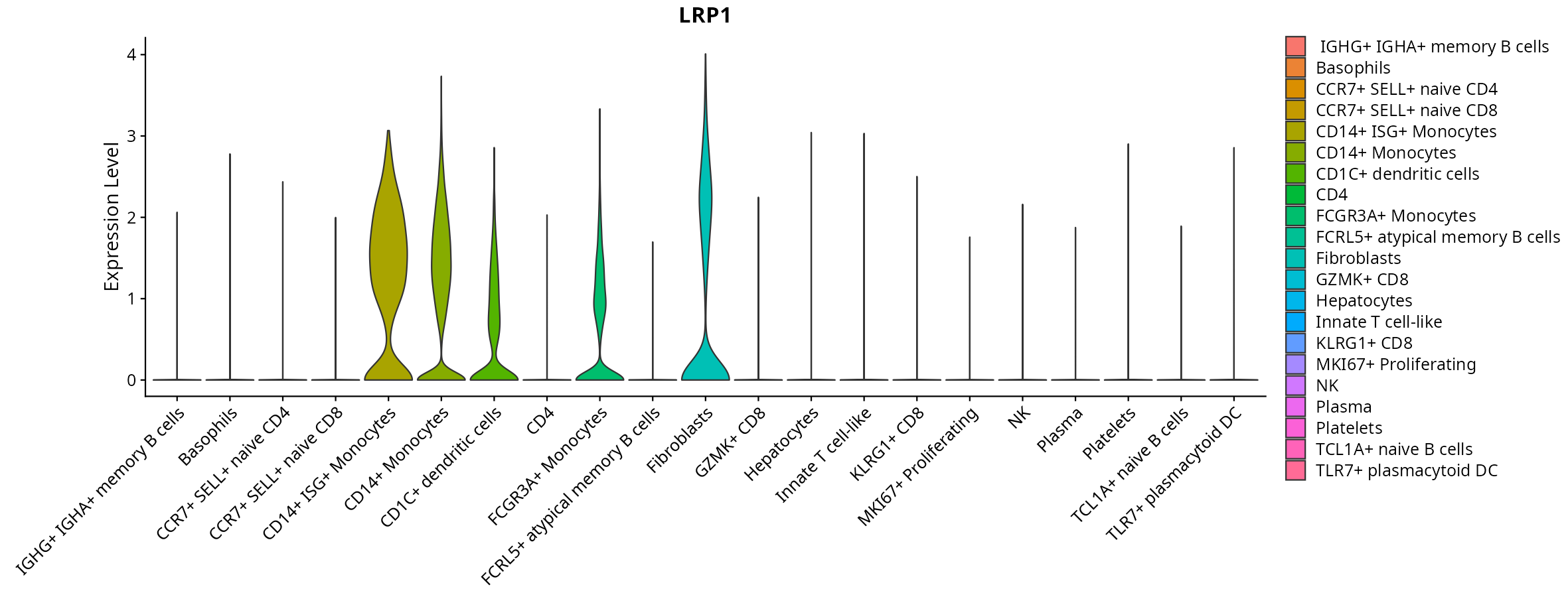
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE252863 - Gene expression in cell subsets
|
|
|
|
|
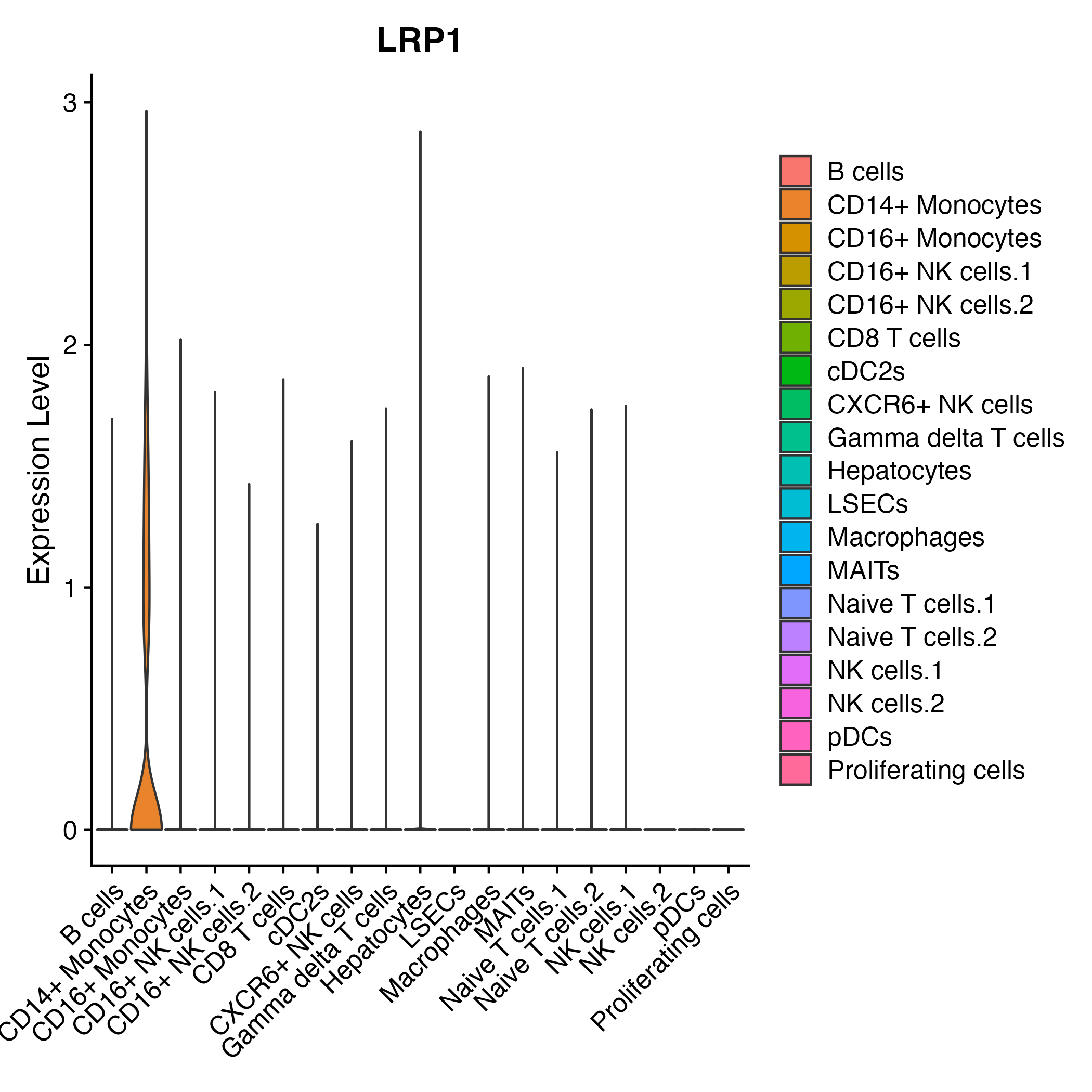
|
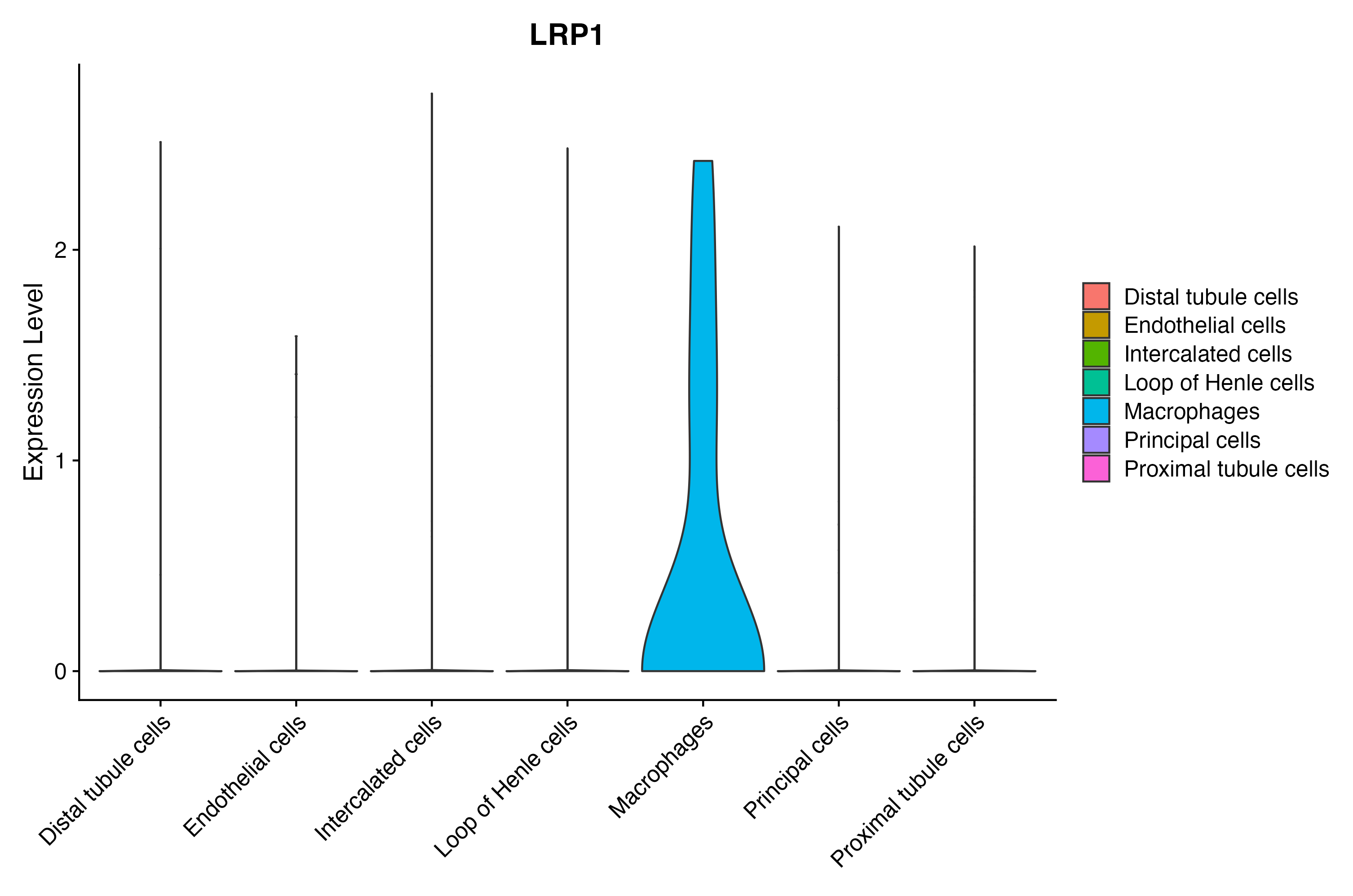
> Dataset: GSE199850 - Gene expression in cell subsets
|
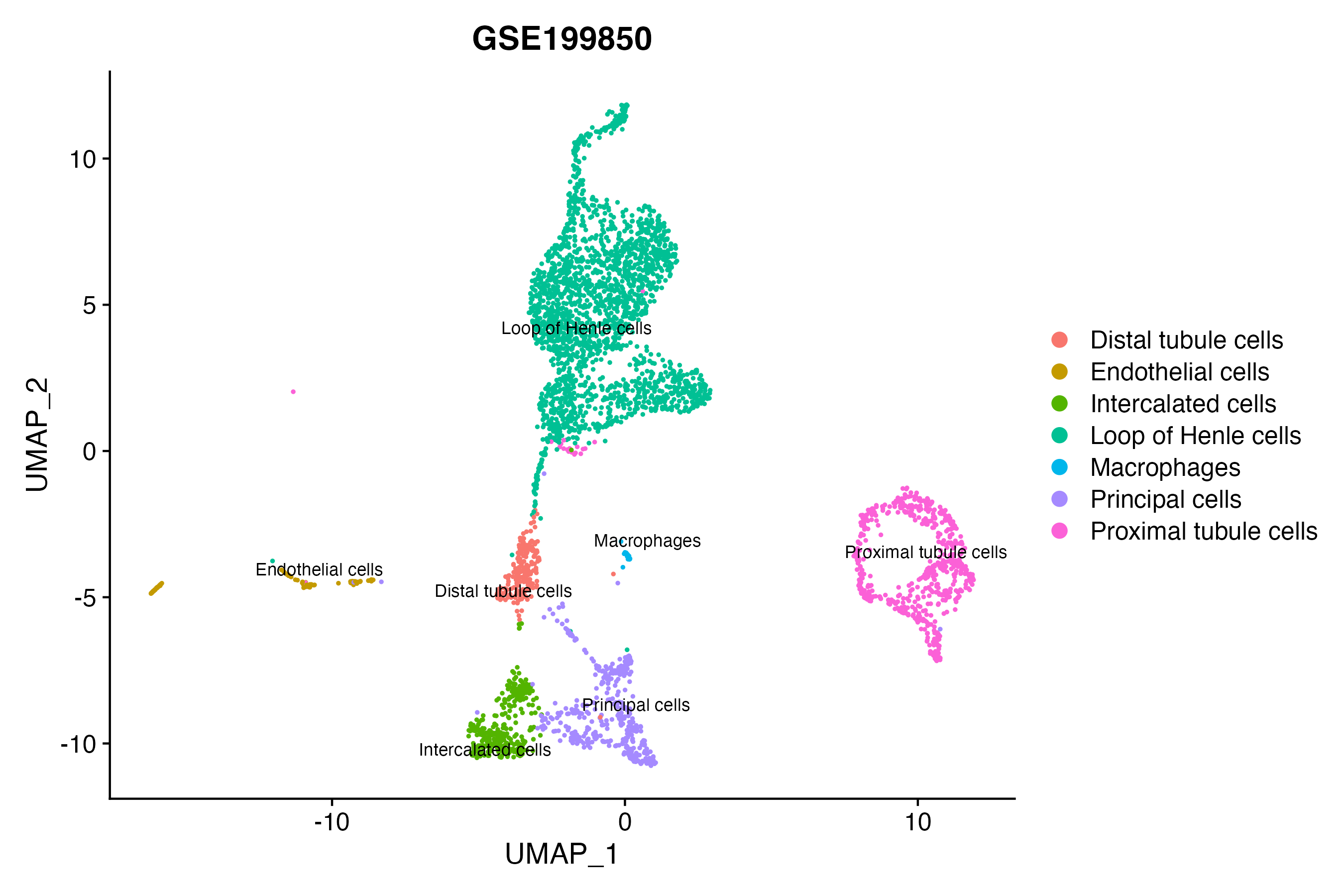
|
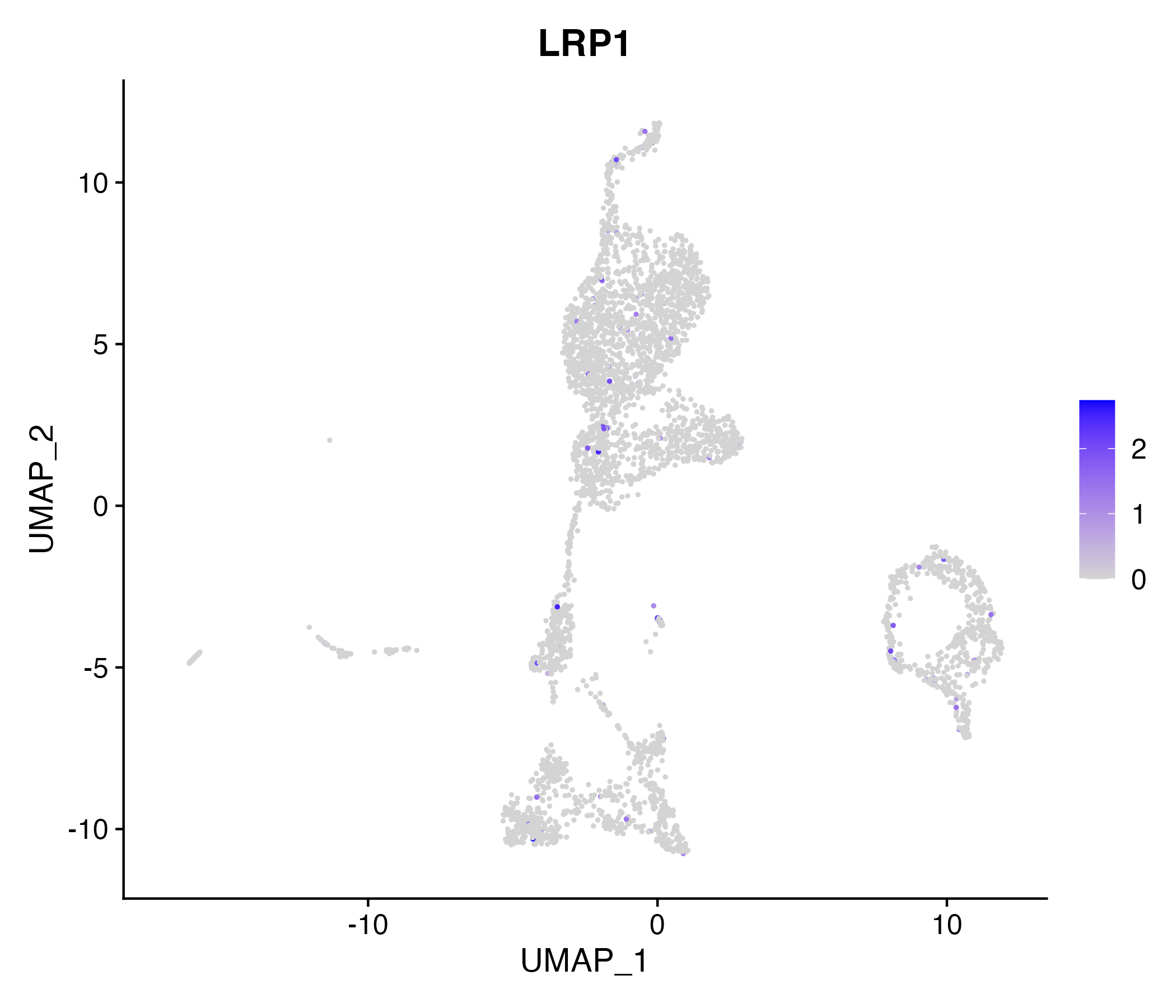
|
|
|
|
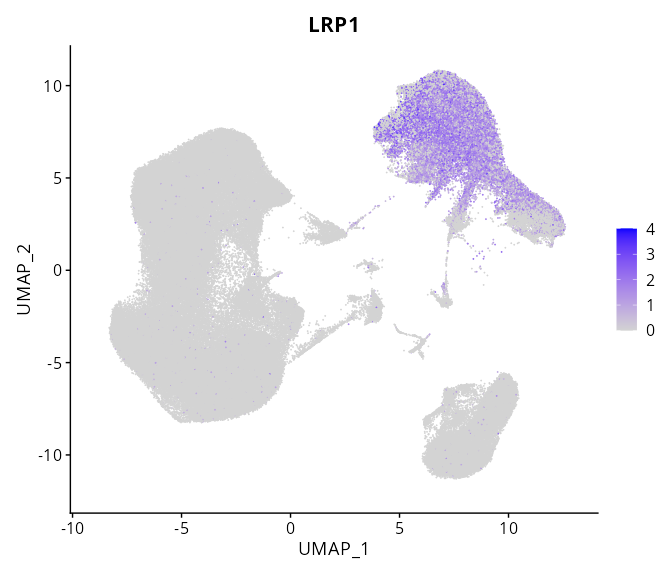
> Dataset: GSE247322 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information