Gene Information
|
Gene Name
|
MAPKAPK3 |
|
Gene ID
|
7867
|
|
Gene Full Name
|
MAPK activated protein kinase 3 |
|
Gene Alias
|
3PK|MAPKAP-K3|MAPKAP3|MAPKAPK-3|MDPT3|MK-3|MK3 |
|
Transcripts
|
ENSG00000114738
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm; 
|
|
Membrane Info
|
Disease related genes, Enzymes, Human disease related genes, Potential drug targets, Predicted intracellular proteins, Predicted membrane proteins |
|
Uniport_ID
|
Q16644
|
|
HGNC ID
|
HGNC:6888
|
|
OMIM ID
|
602130 |
|
Summary
|
This gene encodes a member of the Ser/Thr protein kinase family. This kinase functions as a mitogen-activated protein kinase (MAP kinase)- activated protein kinase. MAP kinases are also known as extracellular signal-regulated kinases (ERKs), act as an integration point for multiple biochemical signals. This kinase was shown to be activated by growth inducers and stress stimulation of cells. In vitro studies demonstrated that ERK, p38 MAP kinase and Jun N-terminal kinase were all able to phosphorylate and activate this kinase, which suggested the role of this kinase as an integrative element of signaling in both mitogen and stress responses. This kinase was reported to interact with, phosphorylate and repress the activity of E47, which is a basic helix-loop-helix transcription factor known to be involved in the regulation of tissue-specific gene expression and cell differentiation. Alternate splicing results in multiple transcript variants that encode the same protein. [provided by RefSeq, Sep 2011] |
Target gene [MAPKAPK3] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1004792 |
chr3 |
51 |
10 |
8 |
69 |
View |
| 1040924 |
chr3 |
1894 |
410 |
138 |
2442 |
View |
Target gene [MAPKAPK3] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
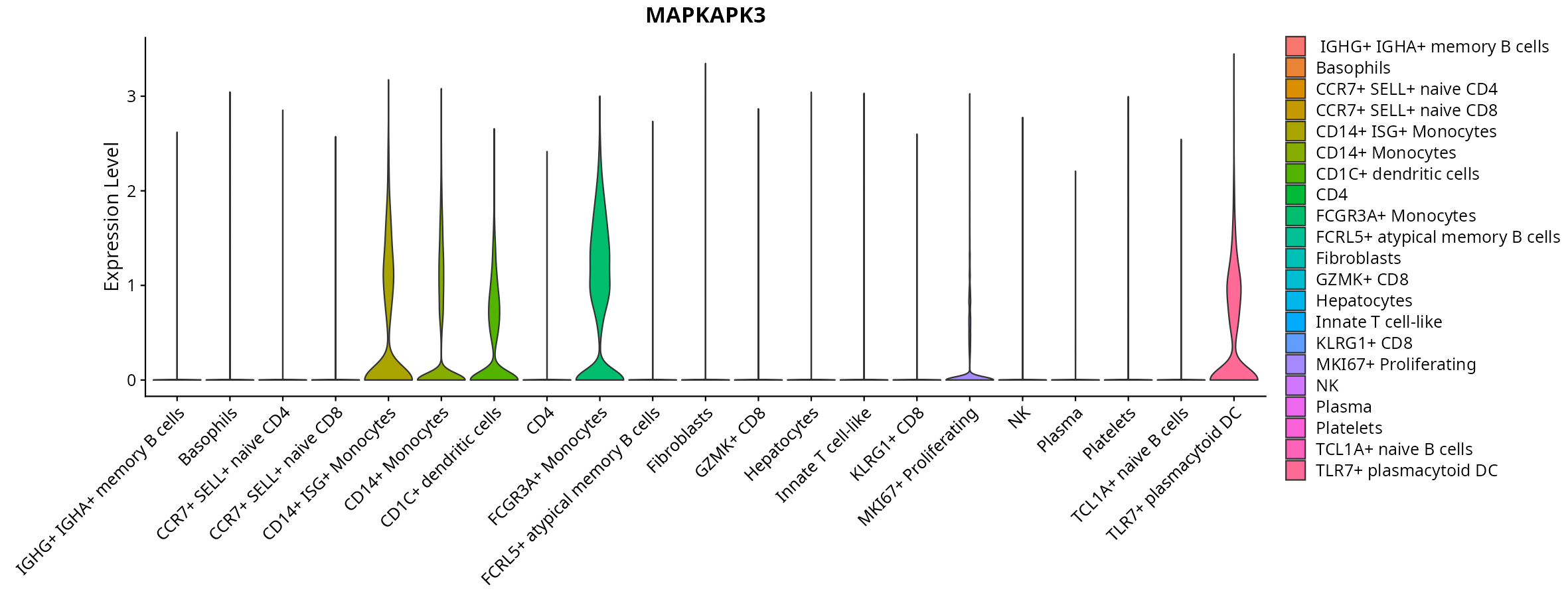
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE252863 - Gene expression in cell subsets
|
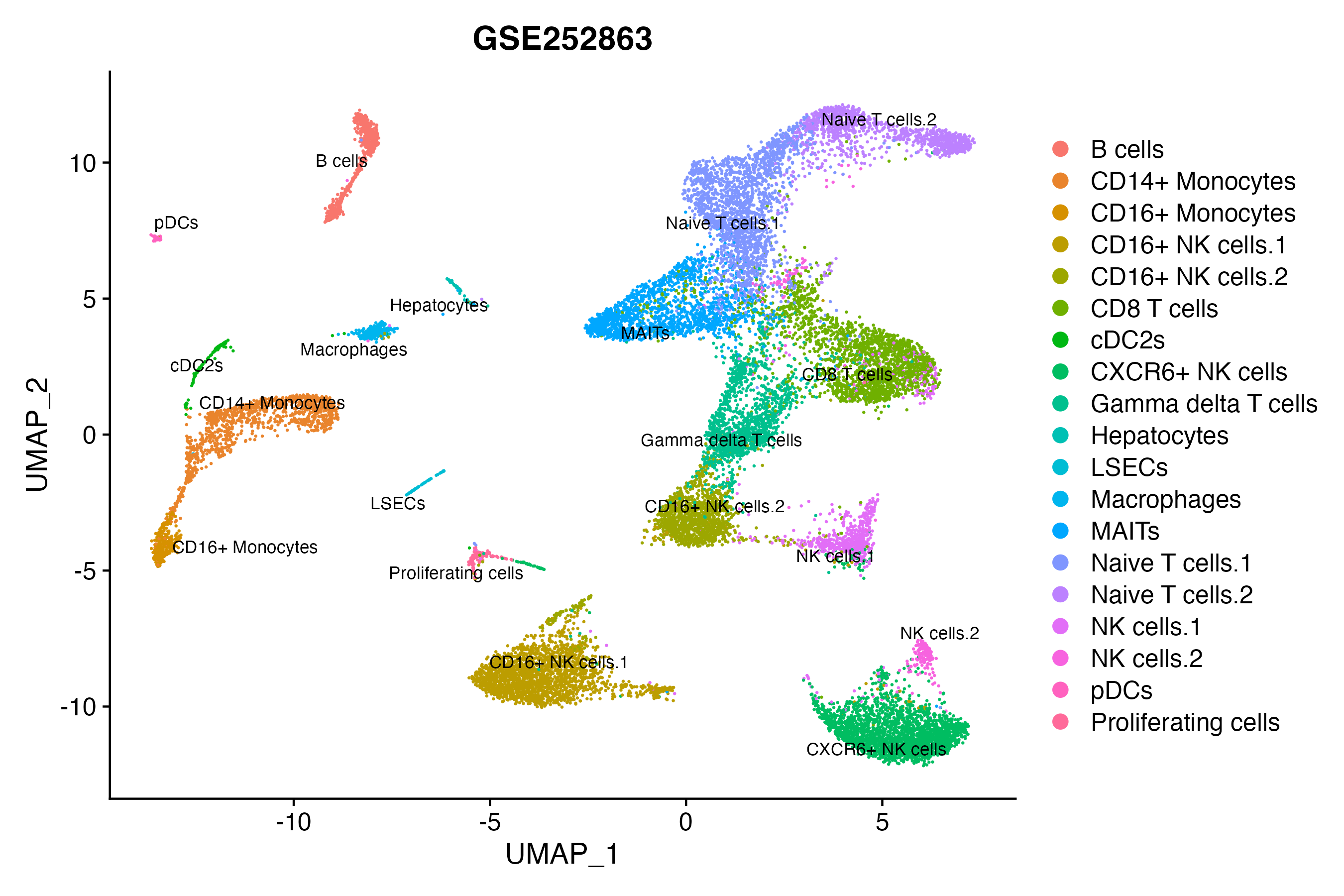
|
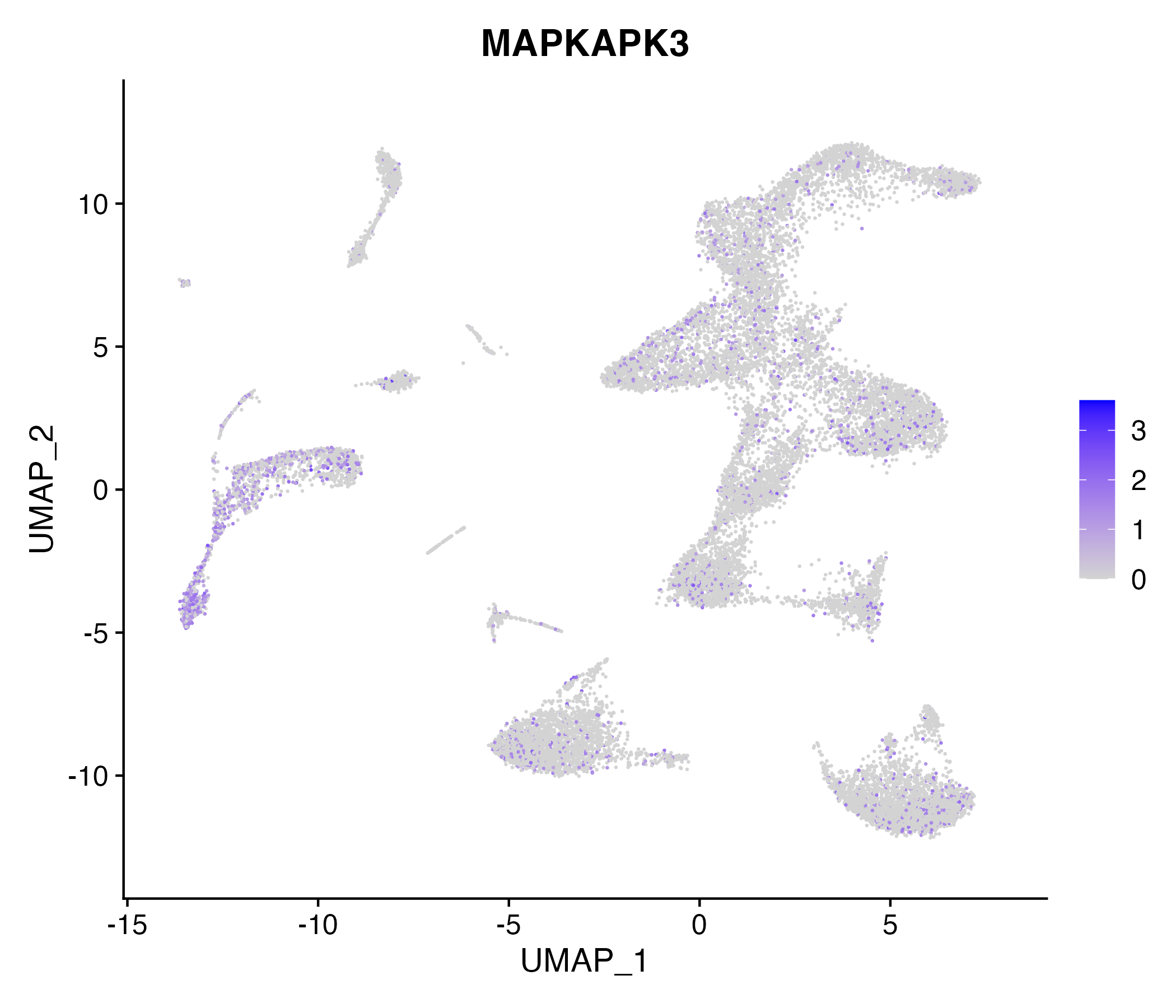
|
|
|
|
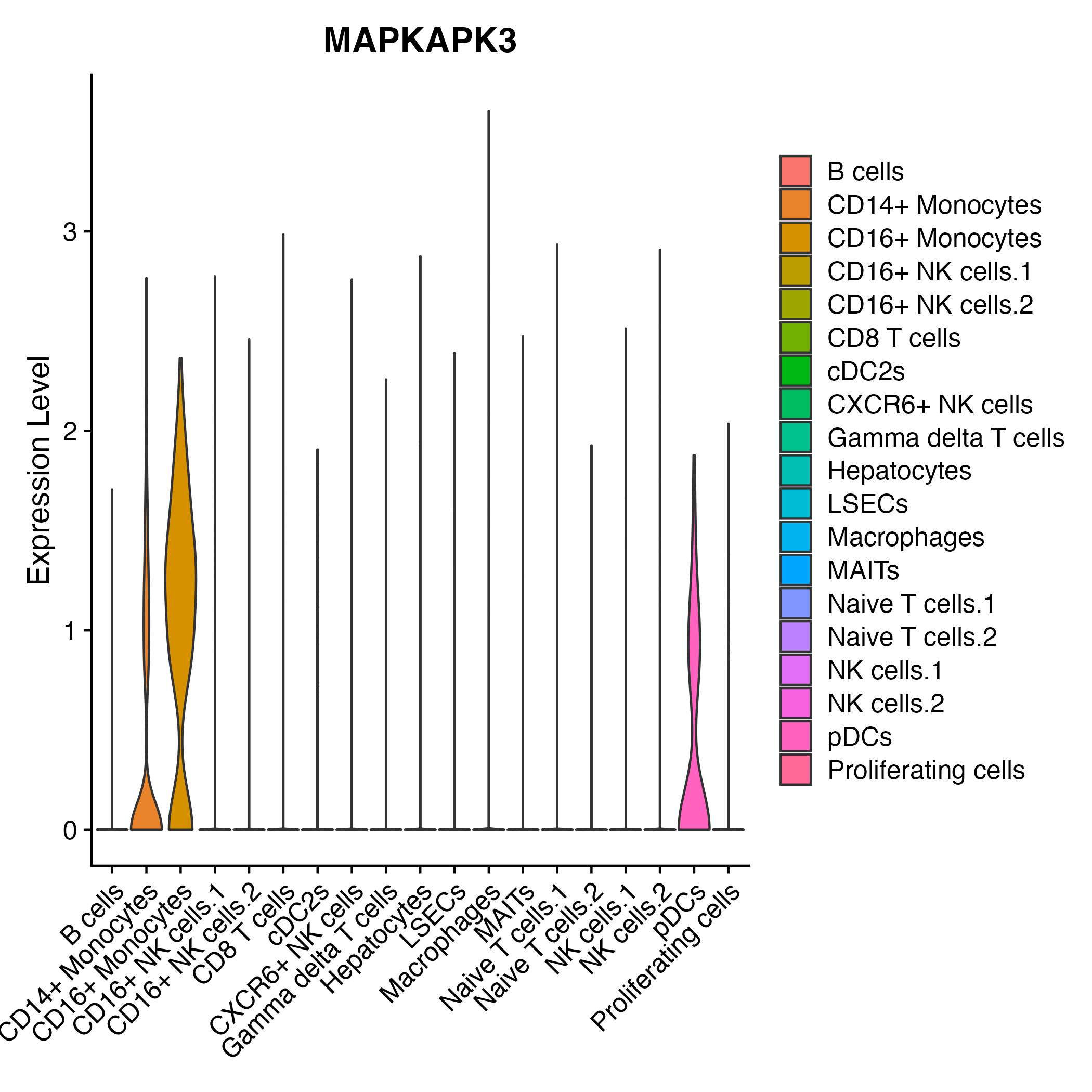
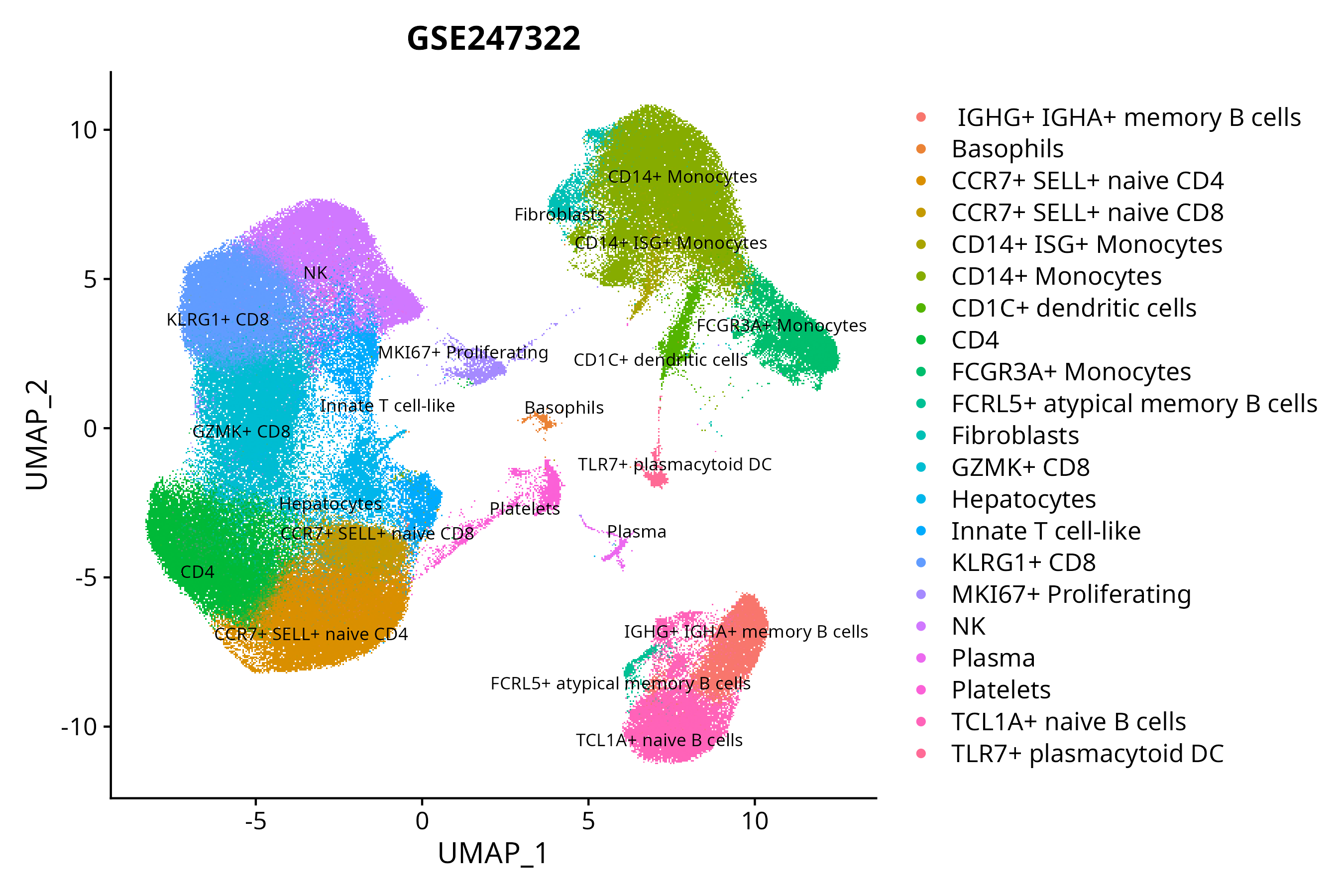
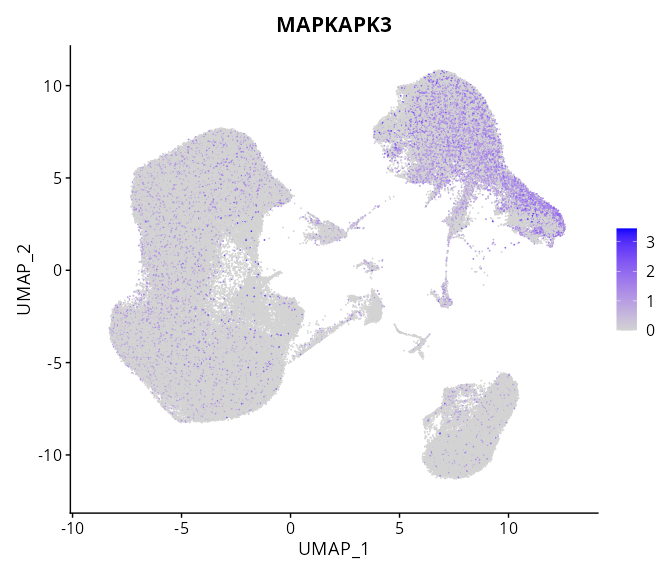
> Dataset: GSE247322 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information