Gene Information
|
Gene Name
|
PRKACA |
|
Gene ID
|
5566
|
|
Gene Full Name
|
protein kinase cAMP-activated catalytic subunit alpha |
|
Gene Alias
|
CAFD1|PKACA|PPNAD4 |
|
Transcripts
|
ENSG00000072062
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Cytosol;Microtubules, Cytokinetic bridge, Primary cilium, Basal body; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Enzymes, Human disease related genes, Metabolic proteins, Potential drug targets, Predicted intracellular proteins, RAS pathway related proteins |
|
Uniport_ID
|
P17612
|
|
HGNC ID
|
HGNC:9380
|
|
OMIM ID
|
601639 |
|
Summary
|
This gene encodes one of the catalytic subunits of protein kinase A, which exists as a tetrameric holoenzyme with two regulatory subunits and two catalytic subunits, in its inactive form. cAMP causes the dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits. Four different regulatory subunits and three catalytic subunits have been identified in humans. cAMP-dependent phosphorylation of proteins by protein kinase A is important to many cellular processes, including differentiation, proliferation, and apoptosis. Constitutive activation of this gene caused either by somatic mutations, or genomic duplications of regions that include this gene, have been associated with hyperplasias and adenomas of the adrenal cortex and are linked to corticotropin-independent Cushing's syndrome. Alternative splicing results in multiple transcript variants encoding different isoforms. Tissue-specific isoforms that differ at the N-terminus have been described, and these isoforms may differ in the post-translational modifications that occur at the N-terminus of some isoforms. [provided by RefSeq, Jan 2015] |
Target gene [PRKACA] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1013246 |
chr19 |
5 |
0 |
0 |
5 |
View |
| 1013247 |
chr19 |
5 |
0 |
0 |
5 |
View |
| 1016399 |
chr19 |
0 |
1 |
0 |
1 |
View |
| 1020873 |
chr19 |
0 |
1 |
0 |
1 |
View |
| 1042353 |
chr19 |
0 |
1 |
0 |
1 |
View |
| 1042354 |
chr19 |
1 |
0 |
0 |
1 |
View |
Target gene [PRKACA] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE68402 - PRKACA peak across samples
|
Peak Plot
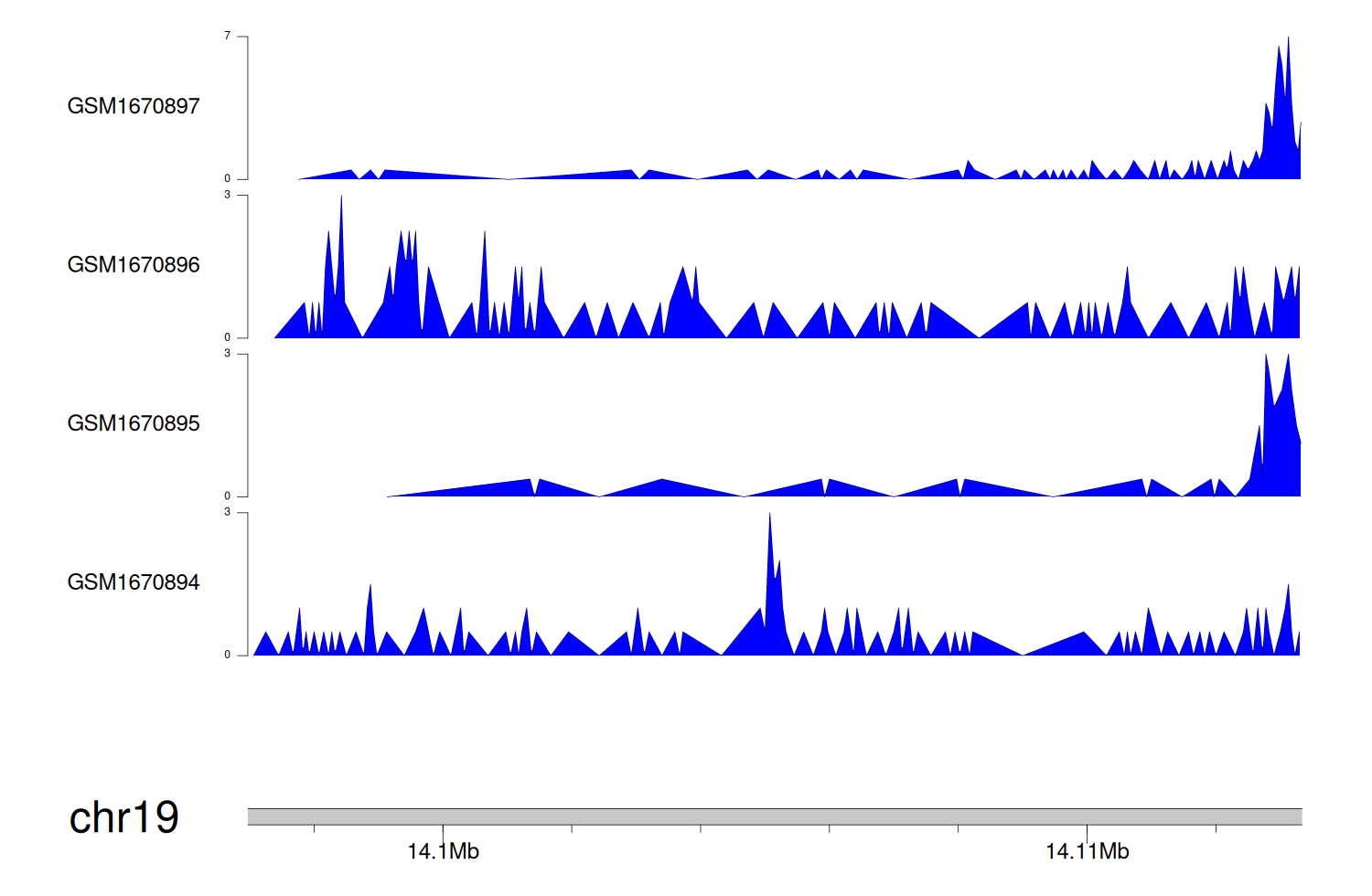
|
> Dataset: GSE270130 - PRKACA peak across samples
|
Peak Plot
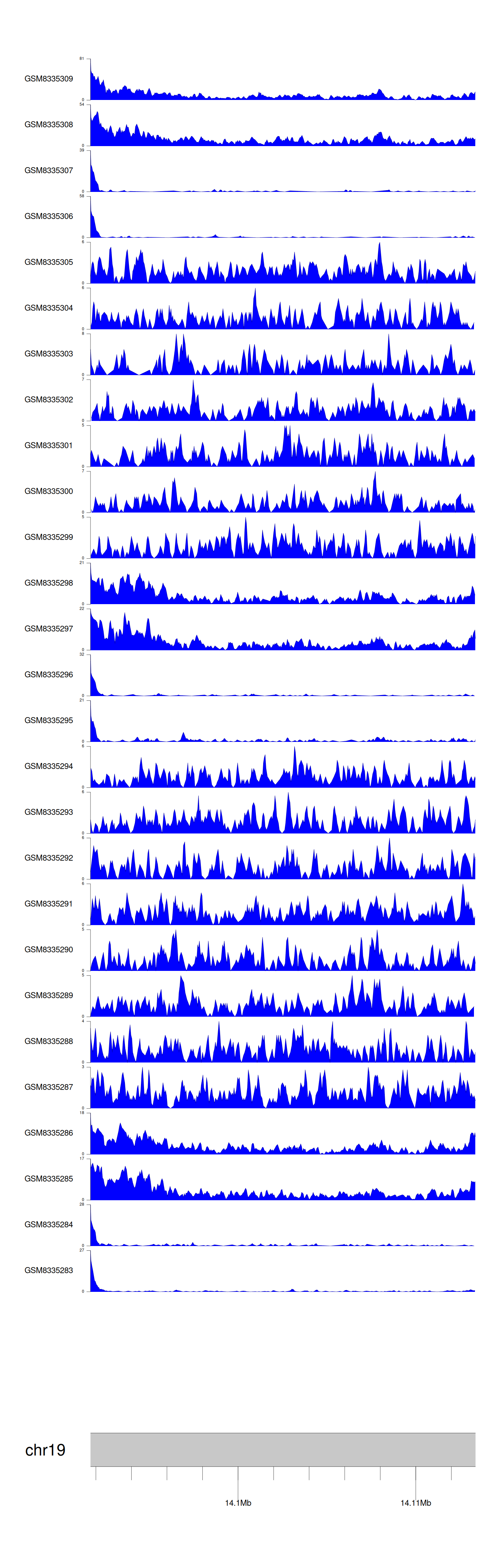
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information