Gene Information
|
Gene Name
|
PTAFR |
|
Gene ID
|
5724
|
|
Gene Full Name
|
platelet activating factor receptor |
|
Gene Alias
|
PAFR |
|
Transcripts
|
ENSG00000169403
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Plasma membrane;Nucleoplasm, Vesicles, Primary cilium; 
|
|
Membrane Info
|
FDA approved drug targets, G-protein coupled receptors, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
P25105
|
|
HGNC ID
|
HGNC:9582
|
|
OMIM ID
|
173393 |
|
Summary
|
This gene encodes a seven-transmembrane G-protein-coupled receptor for platelet-activating factor (PAF) that localizes to lipid rafts and/or caveolae in the cell membrane. PAF (1-0-alkyl-2-acetyl-sn-glycero-3-phosphorylcholine) is a phospholipid that plays a significant role in oncogenic transformation, tumor growth, angiogenesis, metastasis, and pro-inflammatory processes. Binding of PAF to the PAF-receptor (PAFR) stimulates numerous signal transduction pathways including phospholipase C, D, A2, mitogen-activated protein kinases (MAPKs), and the phosphatidylinositol-calcium second messenger system. Following PAFR activation, cells become rapidly desensitized and this refractory state is dependent on PAFR phosphorylation, internalization, and down-regulation. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2011] |
Target gene [PTAFR] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1008004 |
chr1 |
14 |
8 |
1 |
23 |
View |
Target gene [PTAFR] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
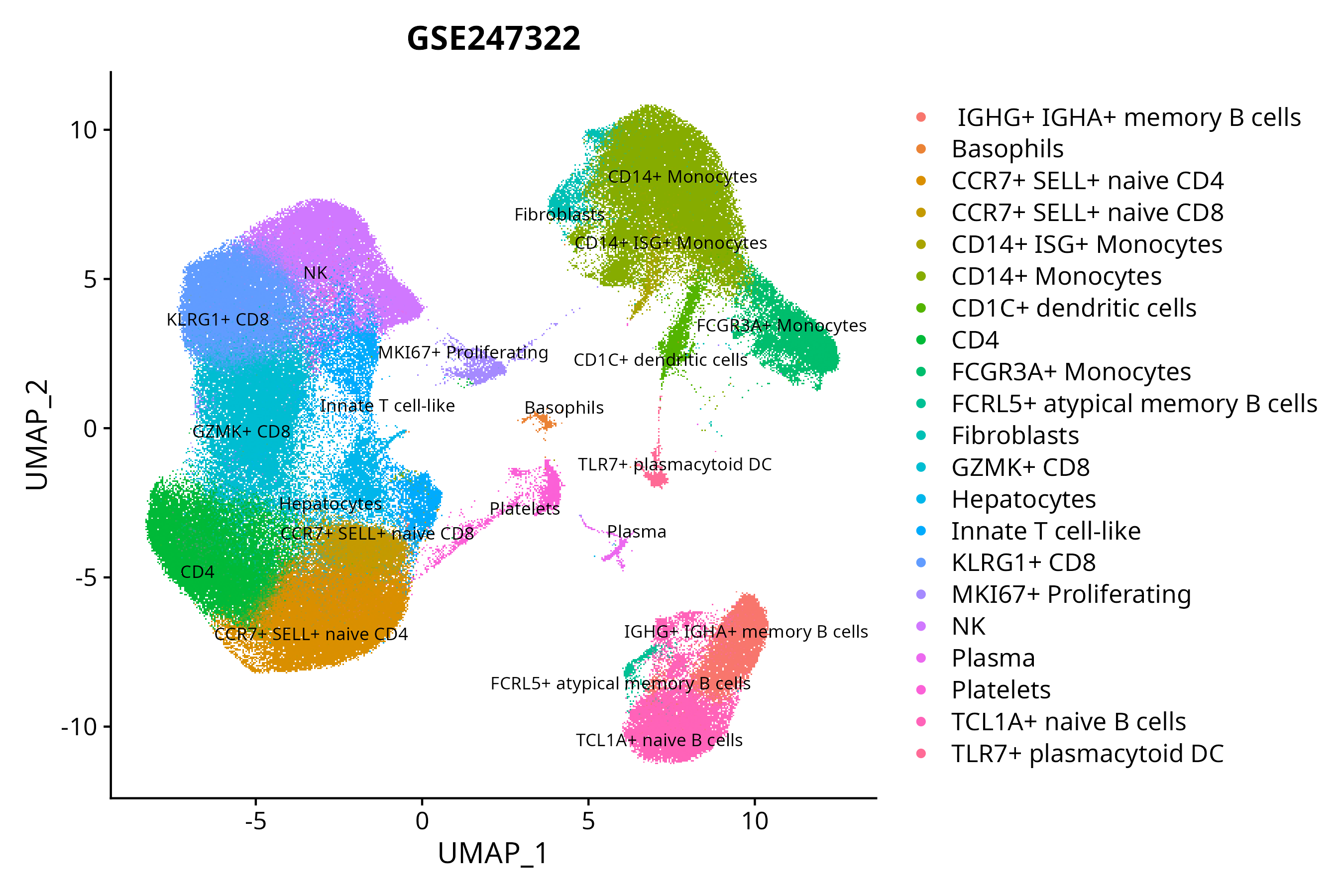
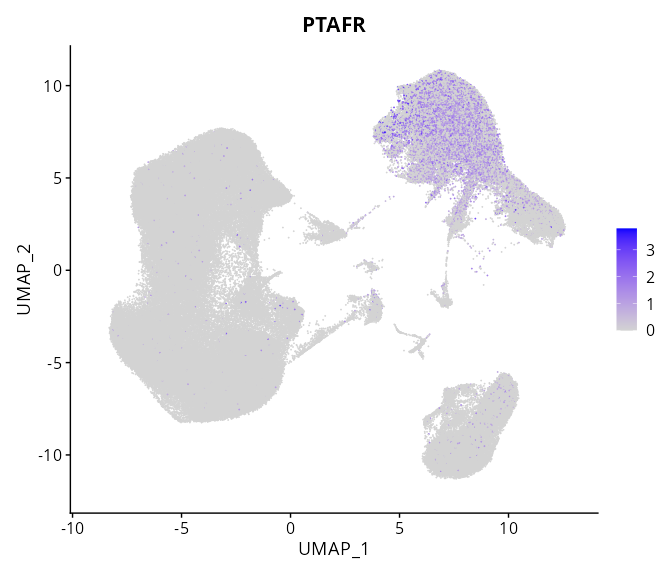
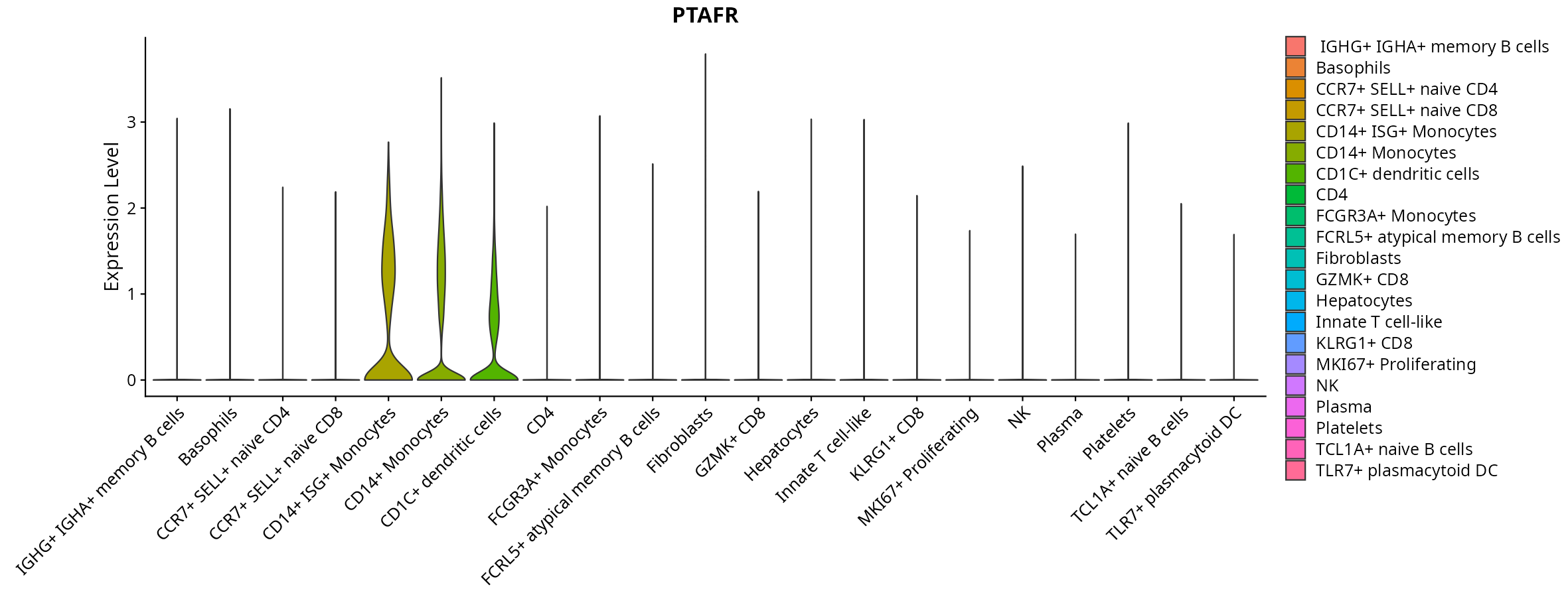
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE247322 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information