Gene Information
|
Gene Name
|
PTPN2 |
|
Gene ID
|
5771
|
|
Gene Full Name
|
protein tyrosine phosphatase non-receptor type 2 |
|
Gene Alias
|
PTN2|PTPT|TC-PTP|TC45|TC48|TCELLPTP|TCPTP |
|
Transcripts
|
ENSG00000175354
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Cytosol; 
|
|
Membrane Info
|
Enzymes, Human disease related genes, Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
P17706
|
|
HGNC ID
|
HGNC:9650
|
|
OMIM ID
|
176887 |
|
Summary
|
The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. Members of the PTP family share a highly conserved catalytic motif, which is essential for the catalytic activity. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. Epidermal growth factor receptor and the adaptor protein Shc were reported to be substrates of this PTP, which suggested the roles in growth factor mediated cell signaling. Multiple alternatively spliced transcript variants encoding different isoforms have been found. Two highly related but distinctly processed pseudogenes that localize to chromosomes 1 and 13, respectively, have been reported. [provided by RefSeq, May 2011] |
Target gene [PTPN2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1014070 |
chr18 |
3 |
1 |
0 |
4 |
View |
Target gene [PTPN2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - PTPN2 peak across samples
|
Peak Plot
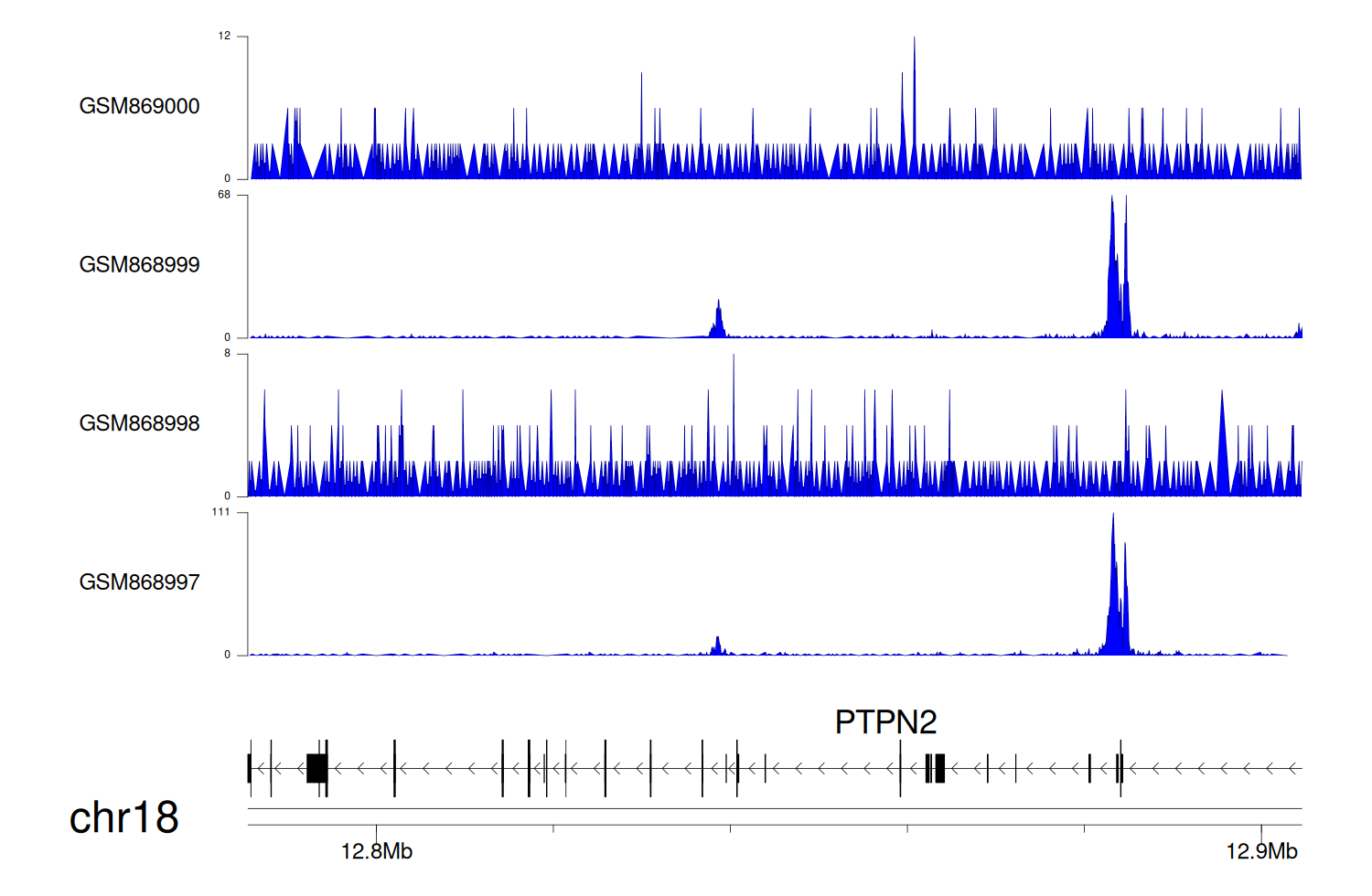
|
> Dataset: GSE68402 - PTPN2 peak across samples
|
Peak Plot
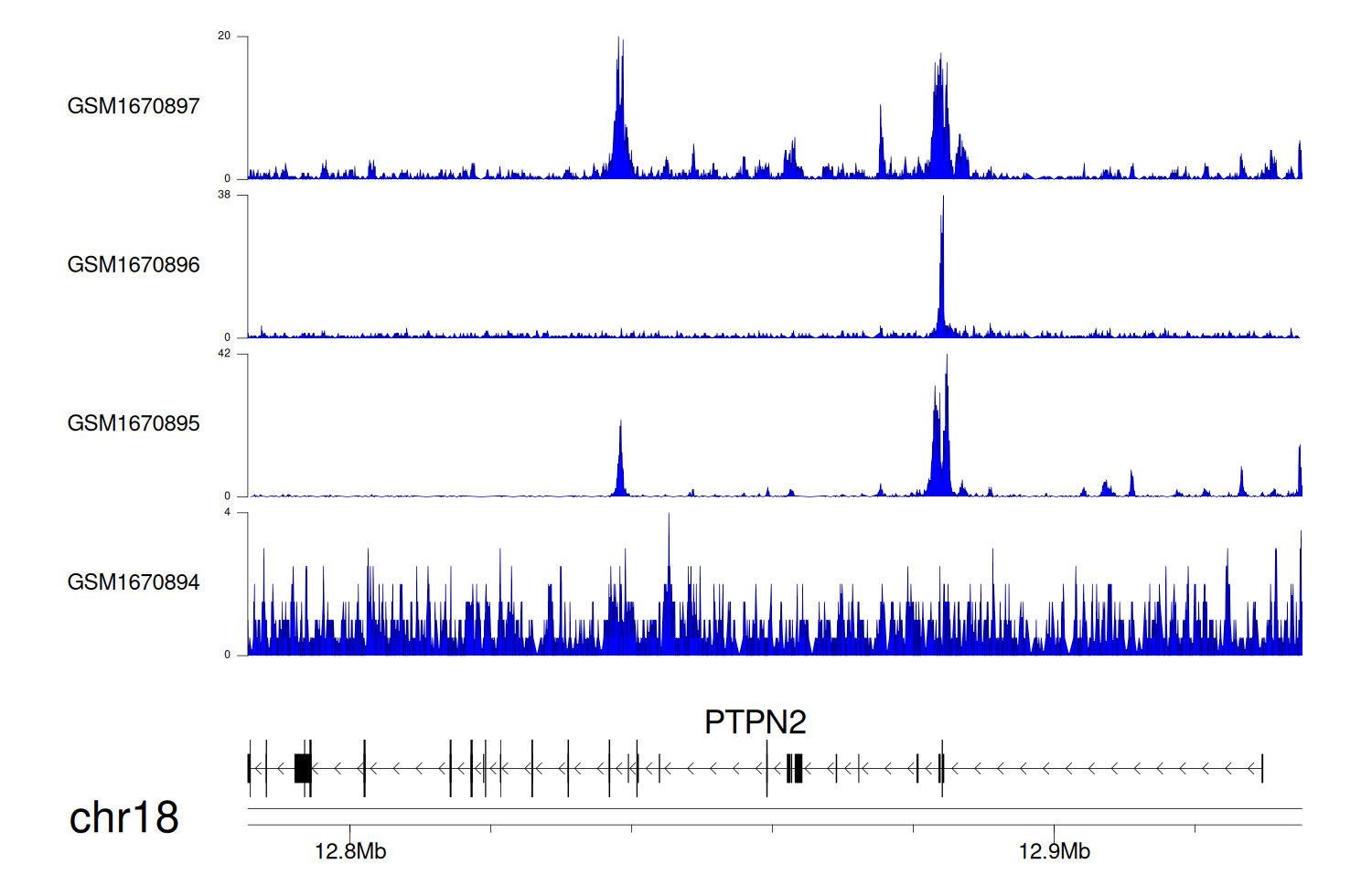
|
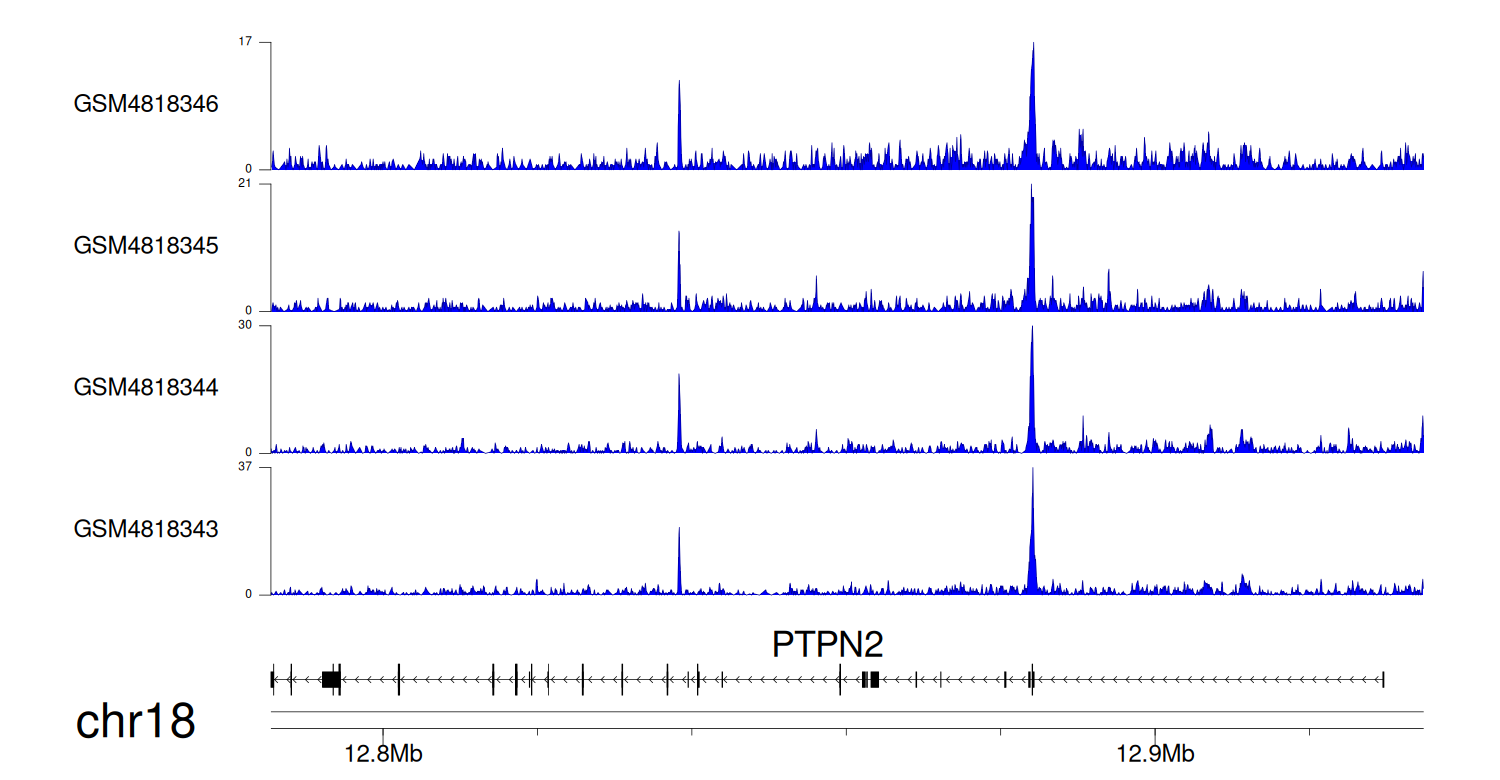
> Dataset: GSE270130 - PTPN2 peak across samples
|
Peak Plot
|
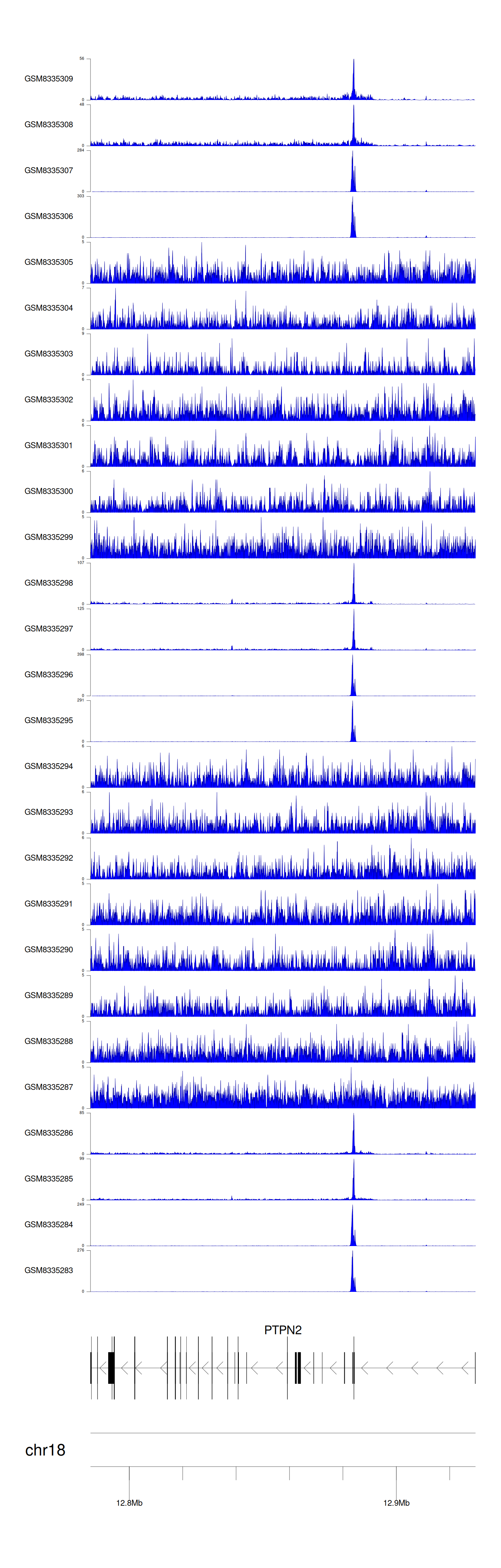
> Dataset: GSE131257 - PTPN2 peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information