Gene Information
|
Gene Name
|
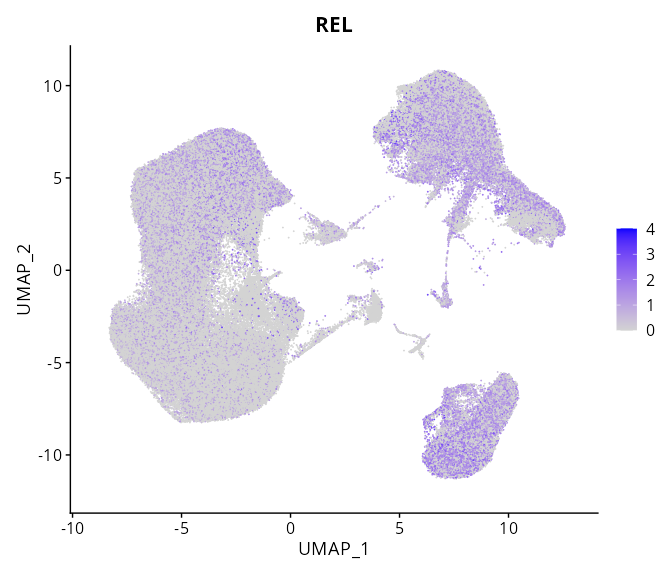
REL |
|
Gene ID
|
5966
|
|
Gene Full Name
|
REL proto-oncogene, NF-kB subunit |
|
Gene Alias
|
C-Rel|HIVEN86A|IMD92 |
|
Transcripts
|
ENSG00000162924
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Predicted intracellular proteins, RAS pathway related proteins, Transcription factors |
|
Uniport_ID
|
Q04864
|
|
HGNC ID
|
HGNC:9954
|
|
OMIM ID
|
164910 |
|
Summary
|
This gene encodes a protein that belongs to the Rel homology domain/immunoglobulin-like fold, plexin, transcription factor (RHD/IPT) family. Members of this family regulate genes involved in apoptosis, inflammation, the immune response, and oncogenic processes. This proto-oncogene plays a role in the survival and proliferation of B lymphocytes. Mutation or amplification of this gene is associated with B-cell lymphomas, including Hodgkin's lymphoma. Single nucleotide polymorphisms in this gene are associated with susceptibility to ulcerative colitis and rheumatoid arthritis. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Apr 2014] |
Target gene [REL] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1003373 |
chr2 |
88 |
46 |
5 |
139 |
View |
Target gene [REL] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
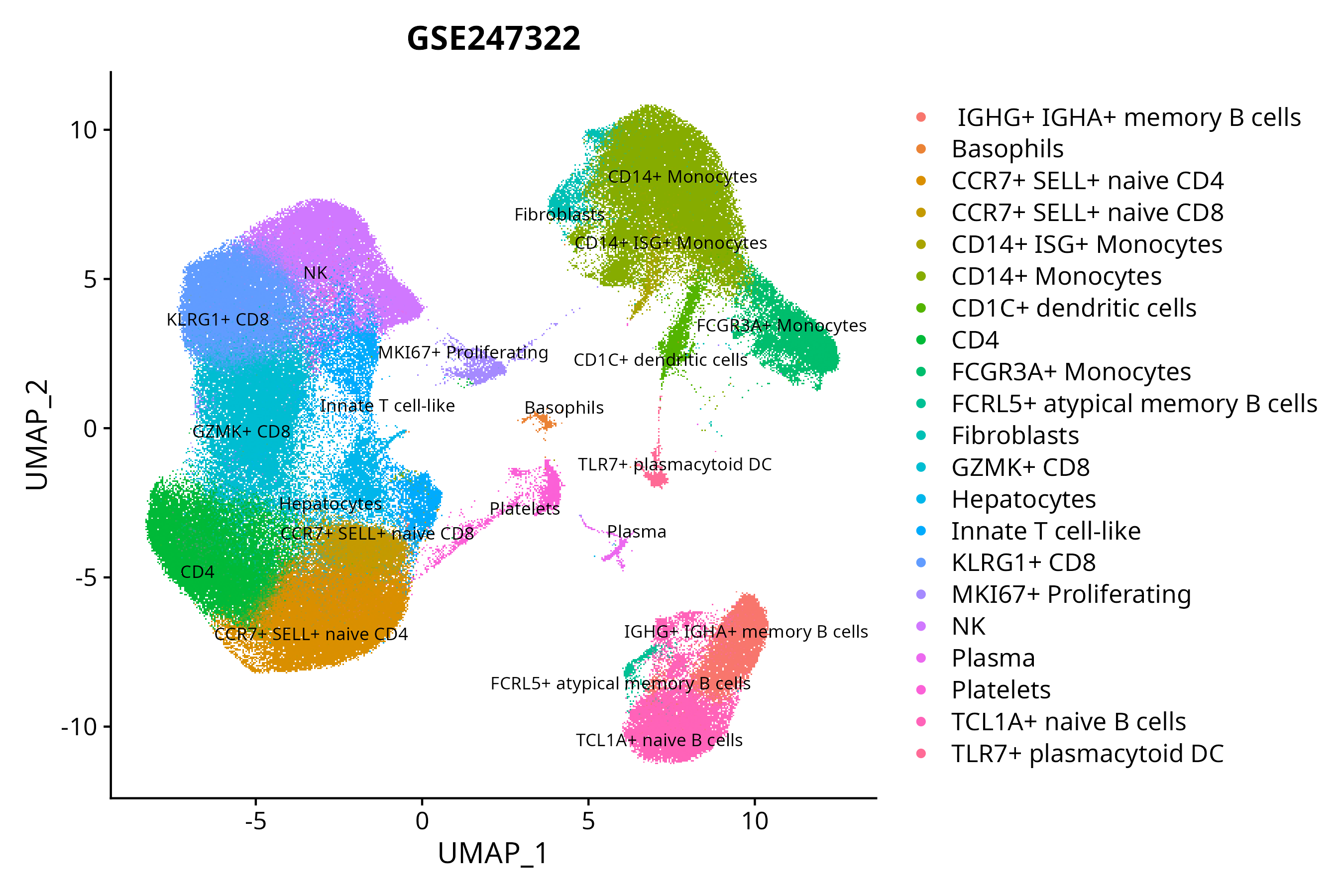
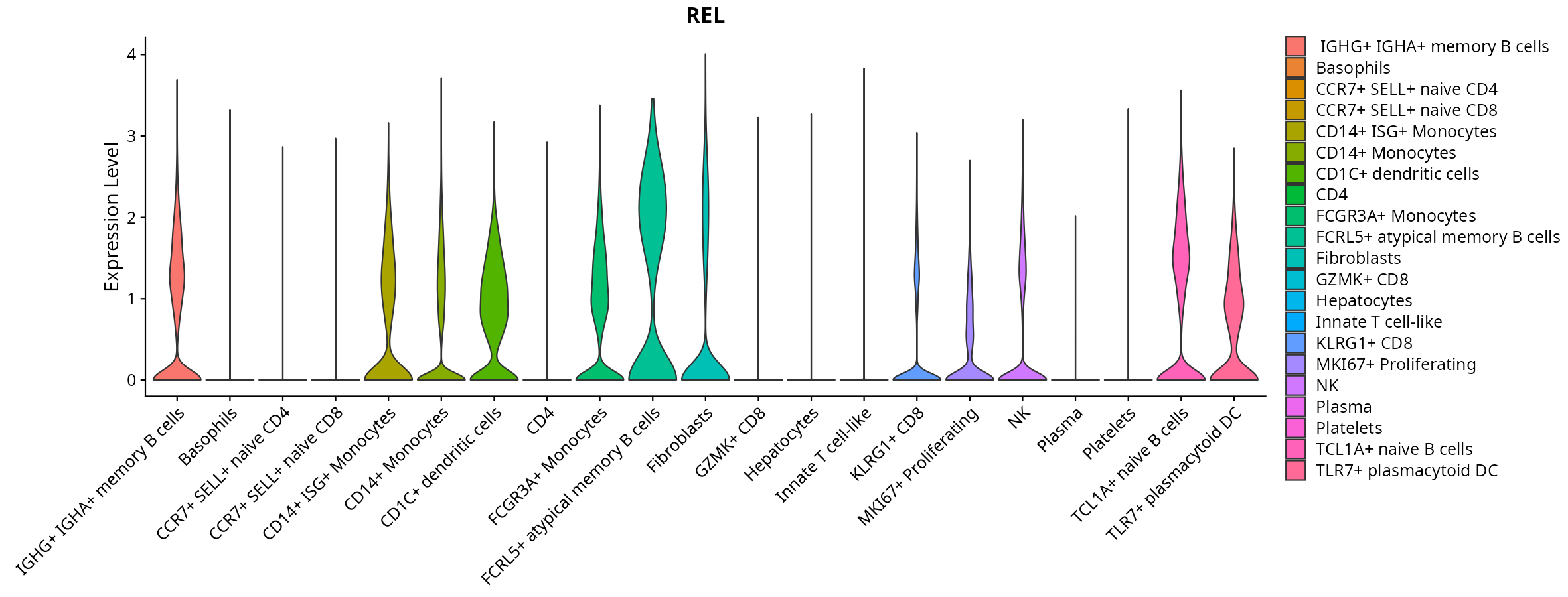
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE247322 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information