Gene Information
|
Gene Name
|
RPA1 |
|
Gene ID
|
6117
|
|
Gene Full Name
|
replication protein A1 |
|
Gene Alias
|
HSSB|MST075|PFBMFT6|REPA1|RF-A|RP-A|RPA70 |
|
Transcripts
|
ENSG00000132383
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm; 
|
|
Membrane Info
|
Disease related genes, Human disease related genes, Plasma proteins, Predicted intracellular proteins |
|
Uniport_ID
|
P27694
|
|
HGNC ID
|
HGNC:10289
|
|
OMIM ID
|
179835 |
|
Summary
|
This gene encodes the largest subunit of the heterotrimeric Replication Protein A (RPA) complex, which binds to single-stranded DNA (ssDNA), forming a nucleoprotein complex that plays an important role in DNA metabolism, being involved in DNA replication, repair, recombination, telomere maintenance, and co-ordinating the cellular response to DNA damage through activation of the ataxia telangiectasia and Rad3-related protein (ATR) kinase. The nucleoprotein complex protects the single-stranded DNA from nucleases, prevents formation of secondary structures that would interfere with repair, and co-ordinates the recruitment and departure of different genome maintenance factors. This subunit contains four oligonucleotide/oligosaccharide-binding (OB) domains, though the majority of ssDNA binding occurs in two of these domains. The heterotrimeric complex has two different modes of ssDNA binding, a low-affinity and high-affinity mode, determined by which ssDNA binding domains are utilized. The different binding modes differ in the length of DNA bound and in the proteins with which it interacts, thereby playing a role in regulating different genomic maintenance pathways. [provided by RefSeq, Sep 2017] |
Target gene [RPA1] related to virus gene/protein/region 
Mutation Table: if previous studies reported that target gene was related to virus gene/region/protein, the information in this literature was listed in this section
| ID |
PMID |
Mutation |
Gene/Protein/Region |
Encoding Gene/Protein |
Disease |
Description |
Detail |
| 1 |
39057060 |
G1862T |
PreC;C
|
C
|
Cell line
|
G
R
S
I
T
|
View |
Target gene [RPA1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1013332 |
chr17 |
5 |
0 |
0 |
5 |
View |
Target gene [RPA1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
TCGA_LIHC_HBV
|
DNA methylation sequencing;RNA-seq |
97 |
TCGA |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
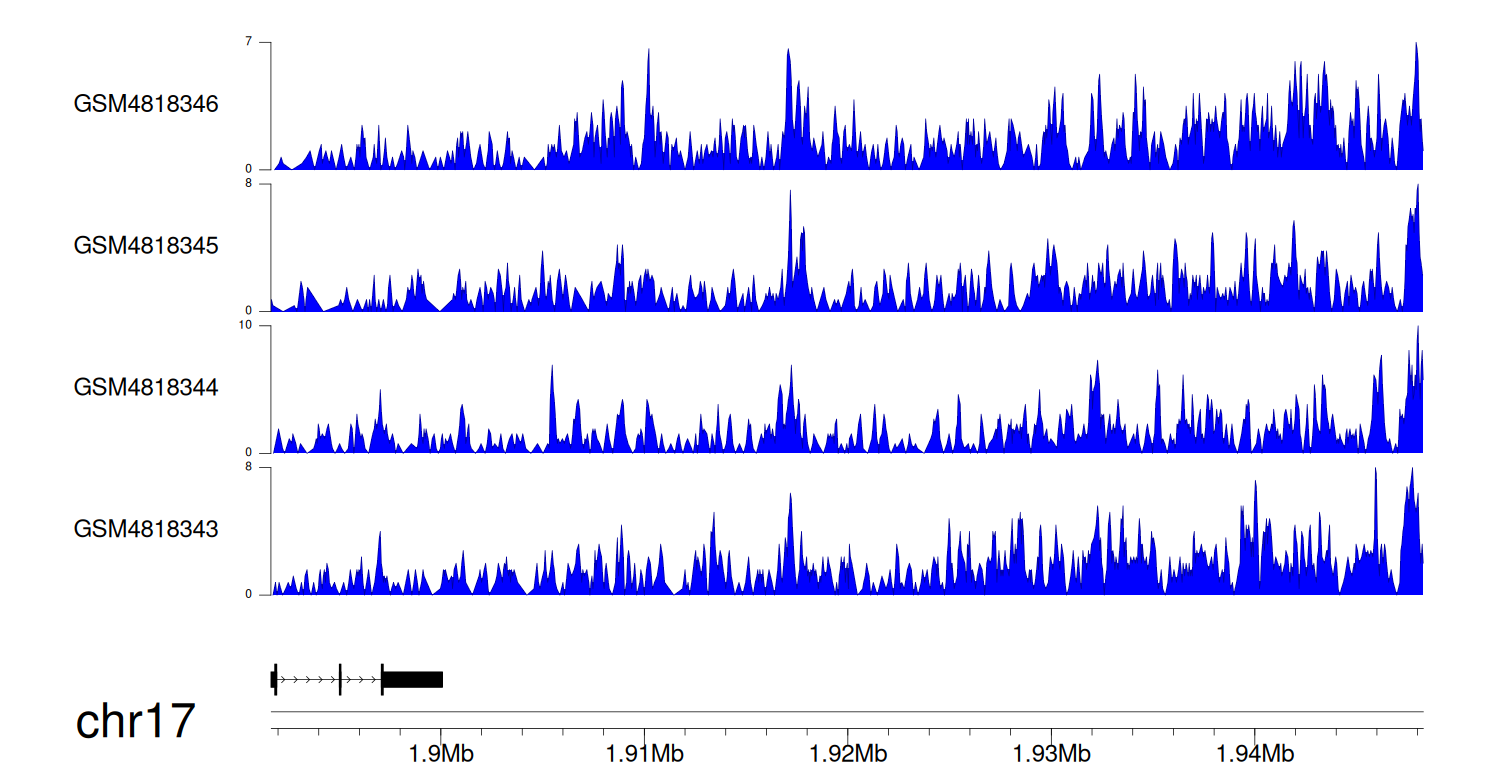
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE35465 - RPA1 peak across samples
|
Peak Plot
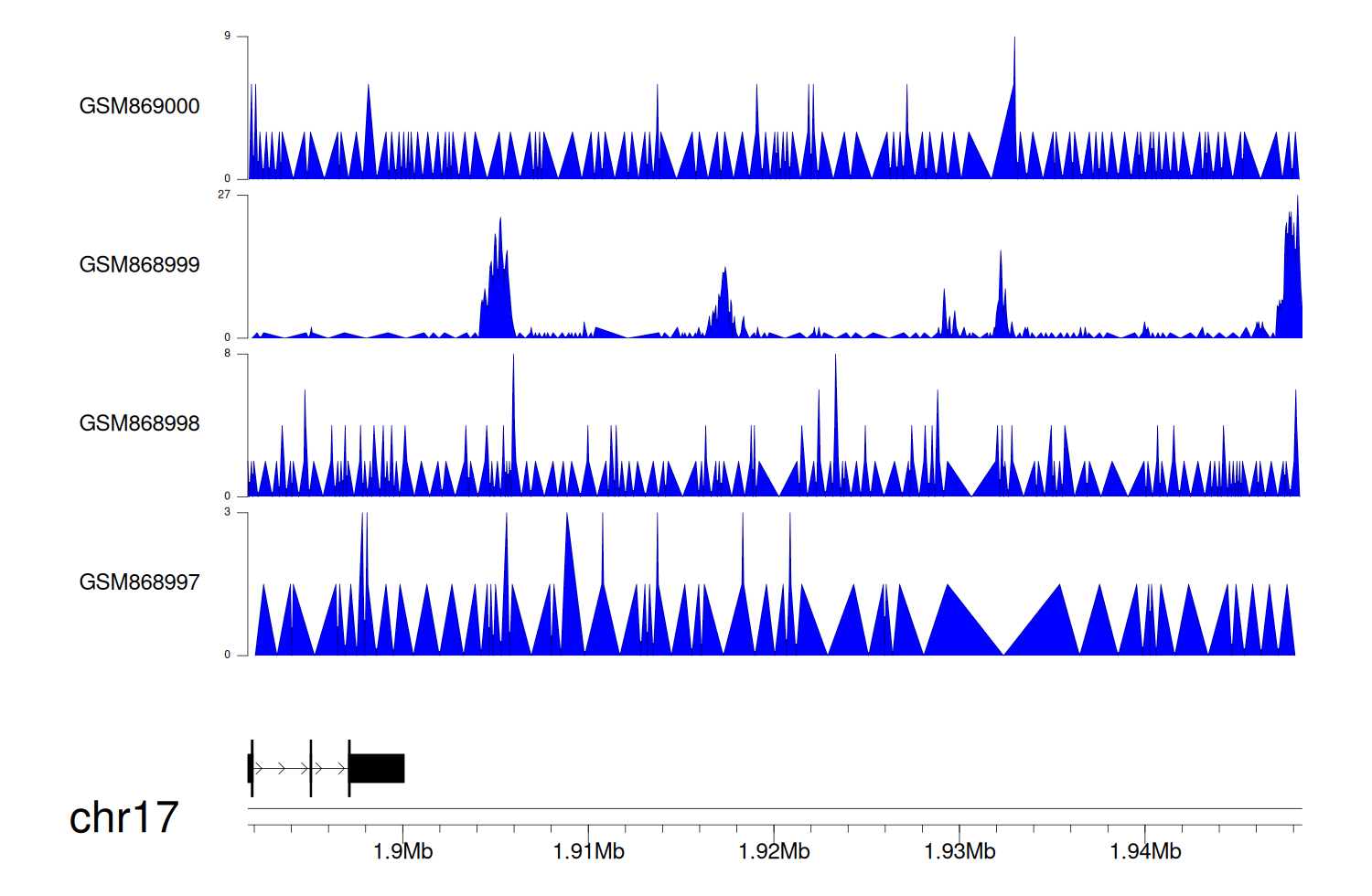
|
> Dataset: GSE68402 - RPA1 peak across samples
|
Peak Plot
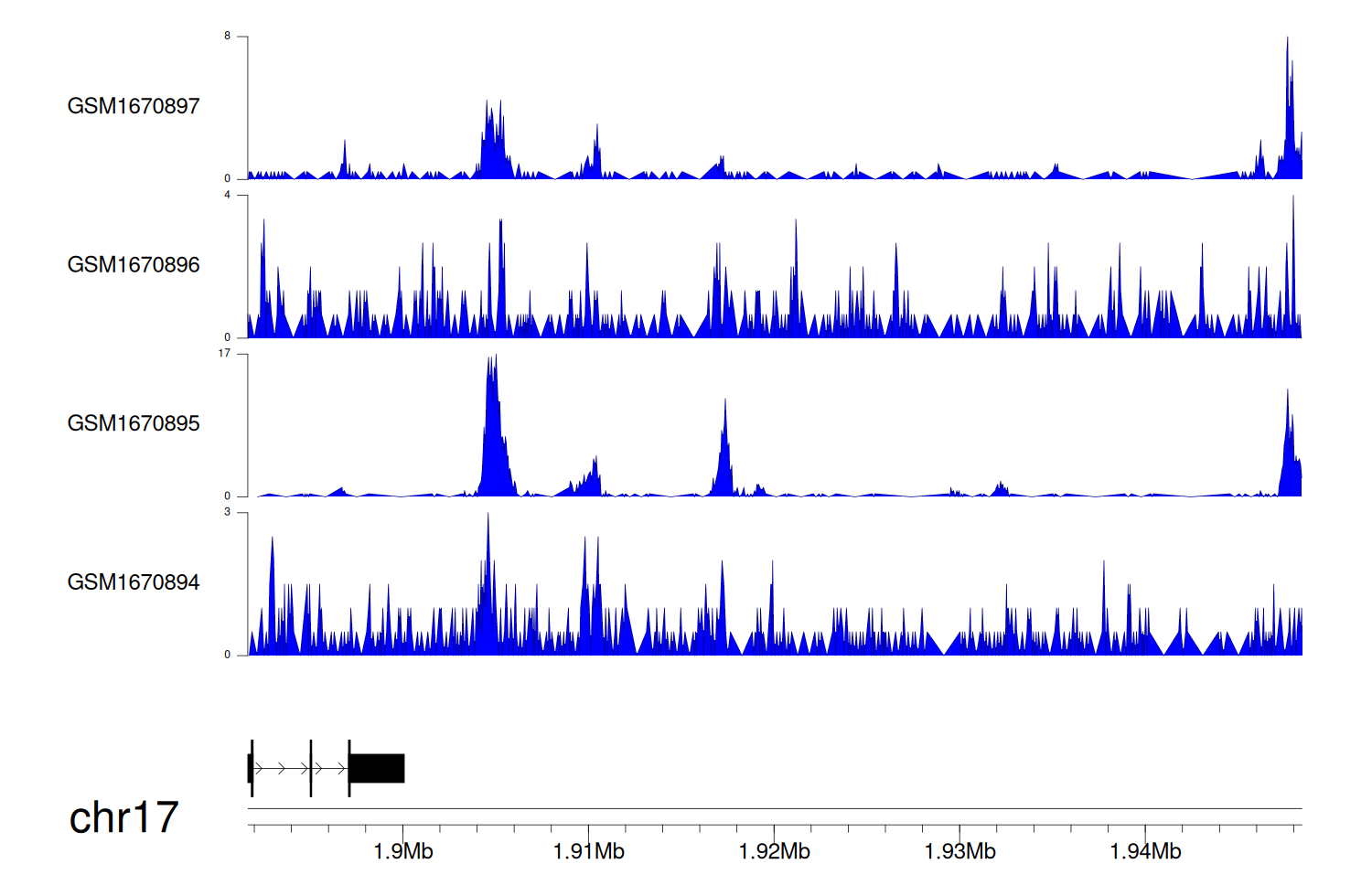
|
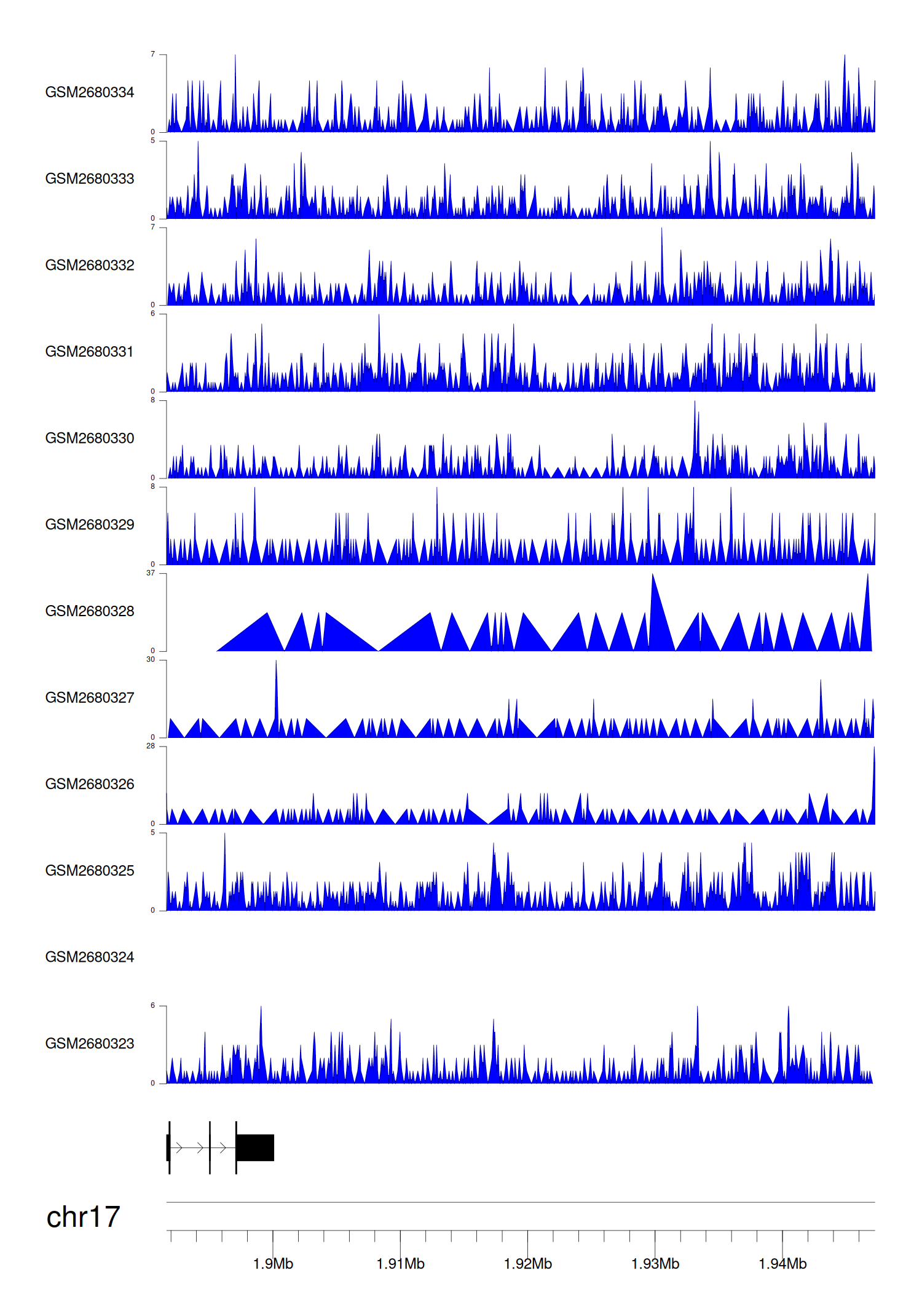
> Dataset: GSE270130 - RPA1 peak across samples
|
Peak Plot
|
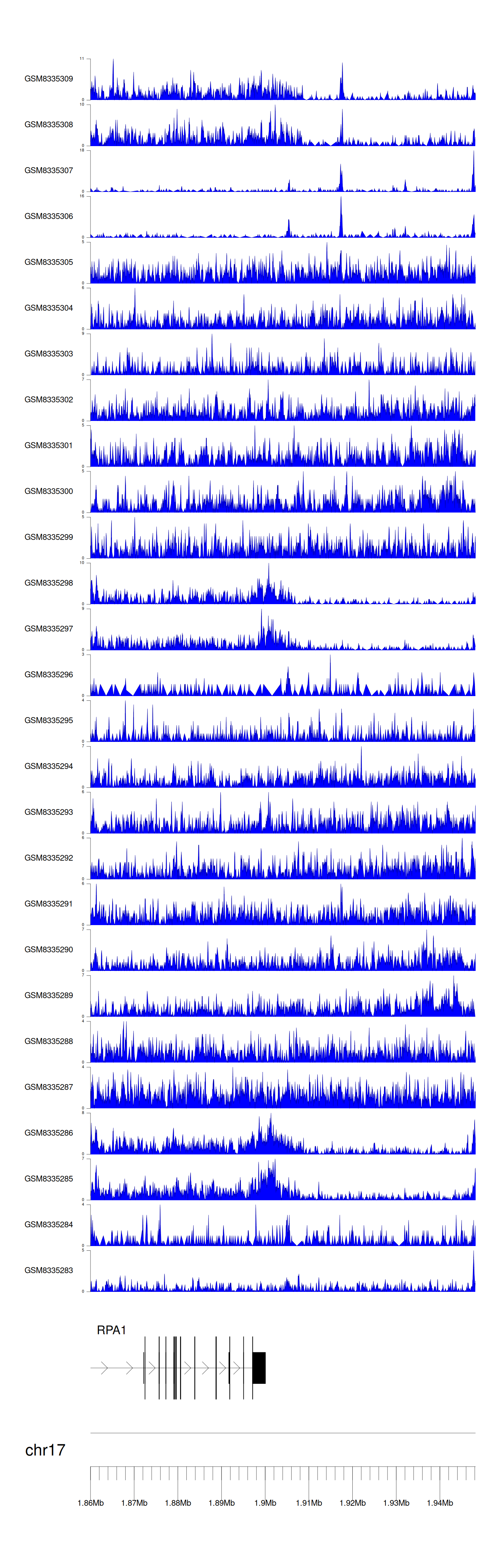
> Dataset: GSE100400 - RPA1 peak across samples
|
Peak Plot
|
> Dataset: GSE131257 - RPA1 peak across samples
|
Peak Plot
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information