Gene Information
|
Gene Name
|
XBP1 |
|
Gene ID
|
7494
|
|
Gene Full Name
|
X-box binding protein 1 |
|
Gene Alias
|
TREB-5|TREB5|XBP-1|XBP2 |
|
Transcripts
|
ENSG00000100219
|
|
Virus
|
HBV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Predicted intracellular proteins, Predicted membrane proteins, Transcription factors |
|
Uniport_ID
|
P17861
|
|
HGNC ID
|
HGNC:12801
|
|
OMIM ID
|
194355 |
|
Summary
|
This gene encodes a transcription factor that regulates MHC class II genes by binding to a promoter element referred to as an X box. This gene product is a bZIP protein, which was also identified as a cellular transcription factor that binds to an enhancer in the promoter of the T cell leukemia virus type 1 promoter. It may increase expression of viral proteins by acting as the DNA binding partner of a viral transactivator. It has been found that upon accumulation of unfolded proteins in the endoplasmic reticulum (ER), the mRNA of this gene is processed to an active form by an unconventional splicing mechanism that is mediated by the endonuclease inositol-requiring enzyme 1 (IRE1). The resulting loss of 26 nt from the spliced mRNA causes a frame-shift and an isoform XBP1(S), which is the functionally active transcription factor. The isoform encoded by the unspliced mRNA, XBP1(U), is constitutively expressed, and thought to function as a negative feedback regulator of XBP1(S), which shuts off transcription of target genes during the recovery phase of ER stress. A pseudogene of XBP1 has been identified and localized to chromosome 5. [provided by RefSeq, Jul 2008] |
Target gene [XBP1] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1003147 |
chr22 |
159 |
2 |
0 |
161 |
View |
Target gene [XBP1] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
S GSE252863
|
scRNA-seq |
10 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE236281
|
RNA-seq |
12 |
Illumina MiSeq (Homo sapiens) |
|
C GSE35465
|
Chip-seq;RNA-seq |
6 |
Illumina HiSeq 2000 (Homo sapiens) |
|
GSE199850
|
scRNA-seq |
1 |
HiSeq X Ten (Homo sapiens) |
|
C GSE68402
|
Chip-seq |
26 |
Illumina MiSeq (Homo sapiens);Illumina HiSeq 2500 (Homo sapiens) |
|
S GSE247322
|
scRNA-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE270130
|
Chip-seq |
27 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE224901
|
RNA-seq |
21 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE100400
|
Chip-seq;RNA-seq;4C_cccDNA |
31 |
Illumina NextSeq 500 (Homo sapiens);Illumina NextSeq 500 (Mus musculus) |
|
GSE173897
|
RNA-seq |
95 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE262515
|
RNA-seq |
21 |
Illumina HiSeq 2500 (Homo sapiens);Illumina HiSeq 2500 (Mus musculus) |
|
GSE110345
|
RNA-seq |
4 |
Illumina HiSeq 2500 (Homo sapiens) |
|
C GSE131257
|
ATAC-seq;RNA-seq |
19 |
Illumina HiSeq 2500 (Homo sapiens) |
|
GSE94660
|
RNA-seq |
42 |
Illumina HiSeq 2500 (Homo sapiens) |
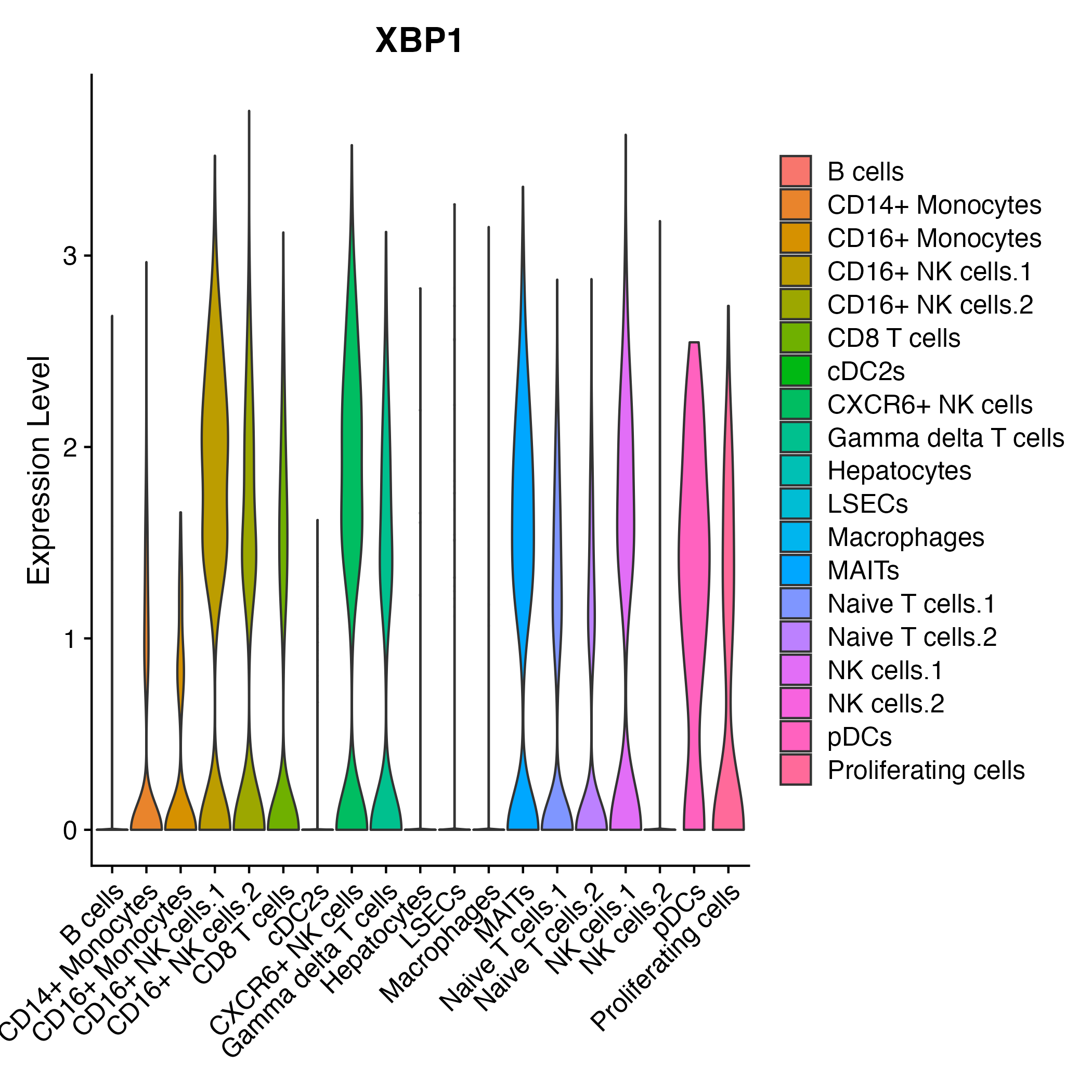
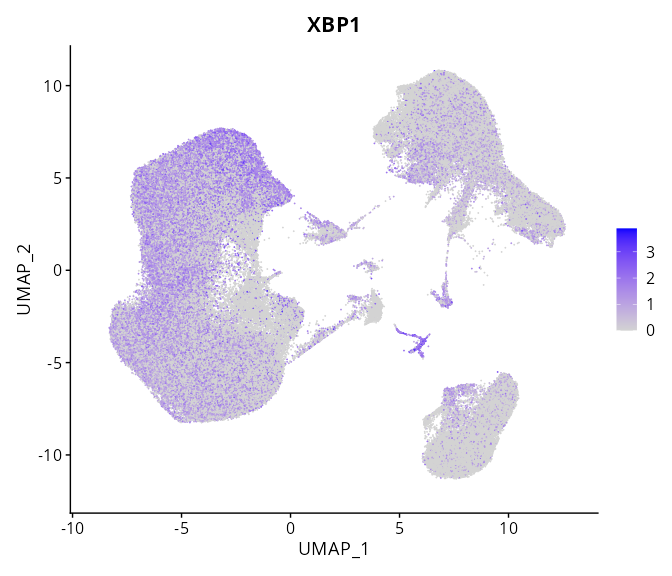
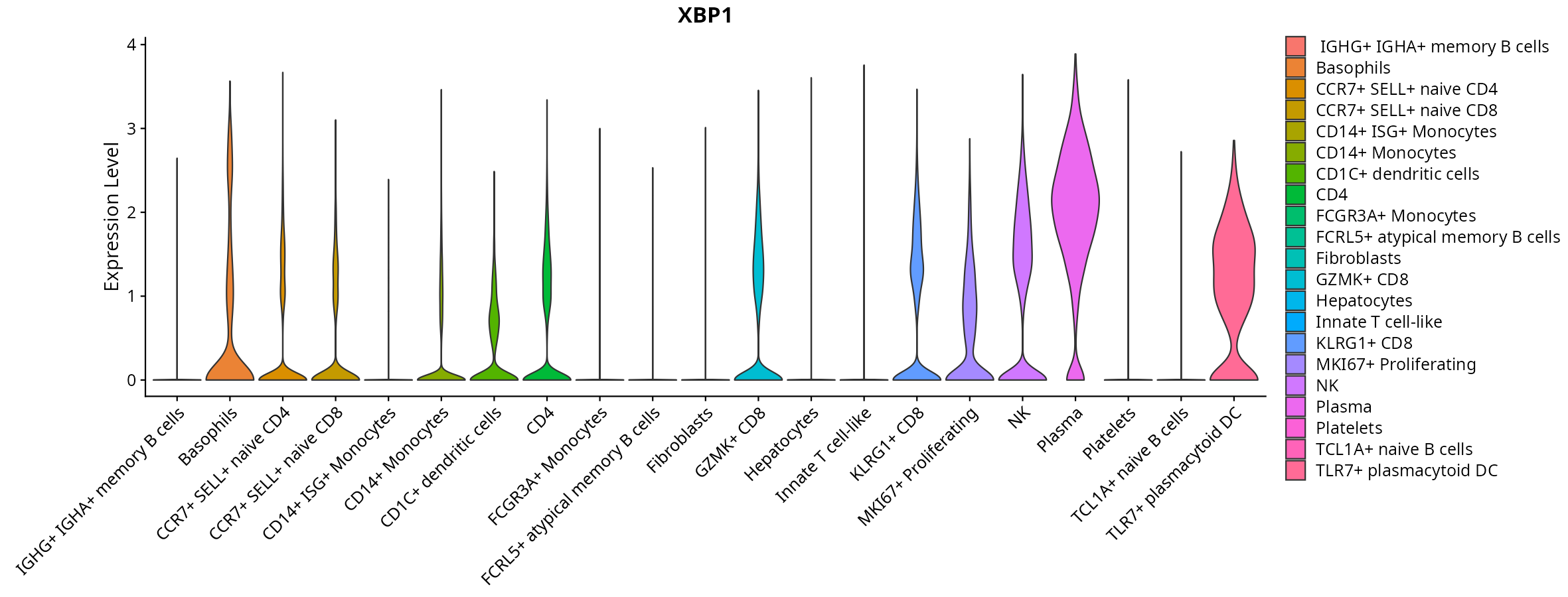
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE252863 - Gene expression in cell subsets
|
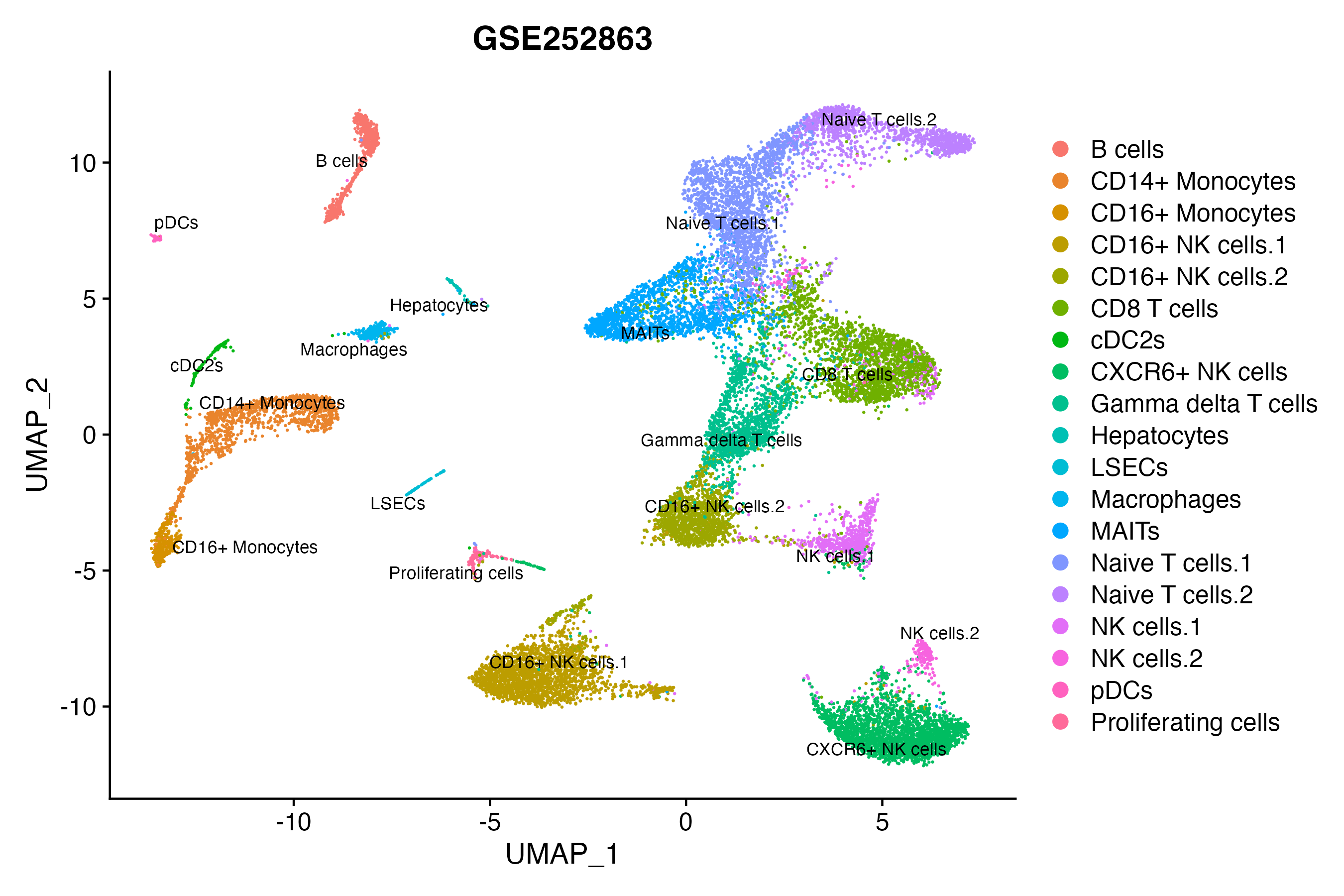
|
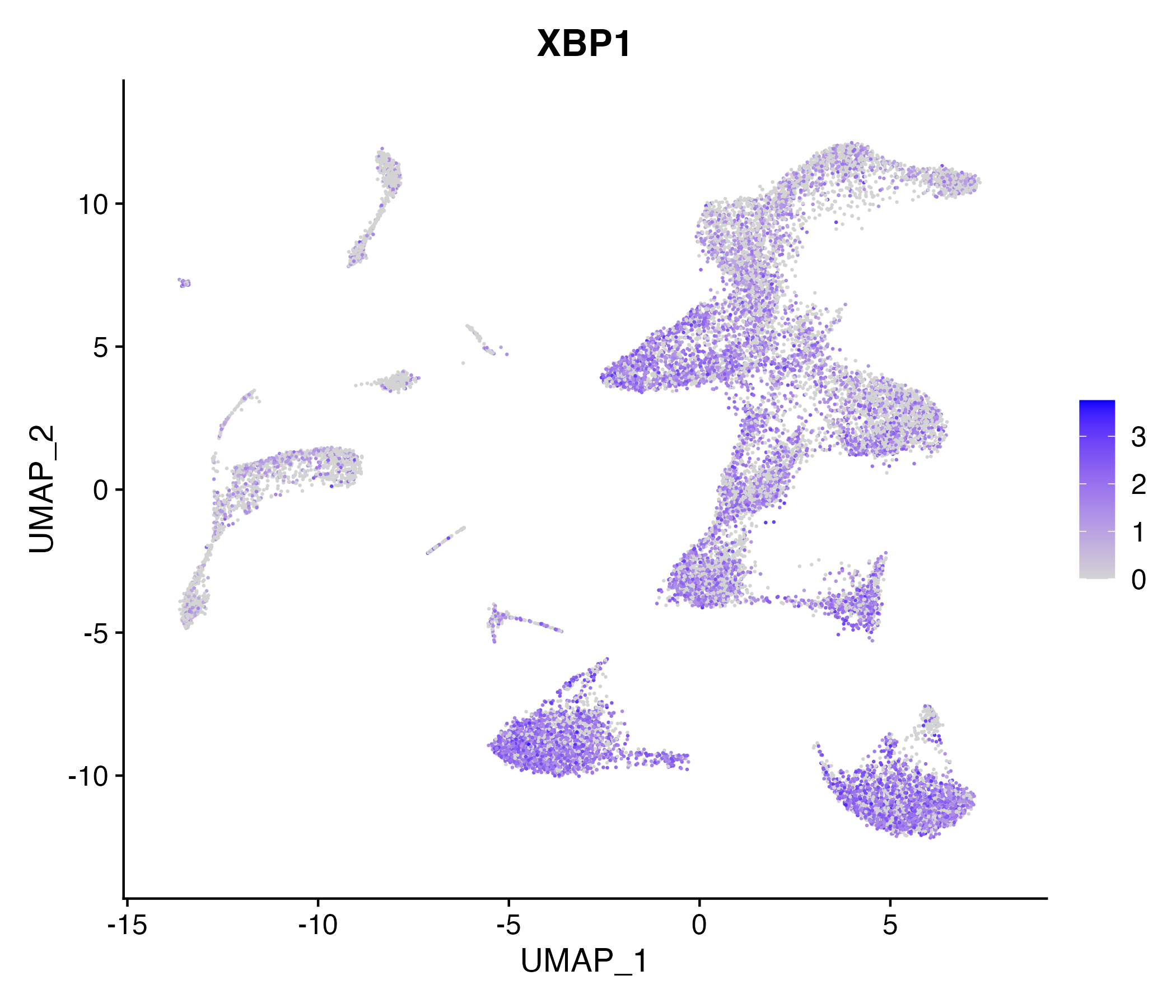
|
|
|
|
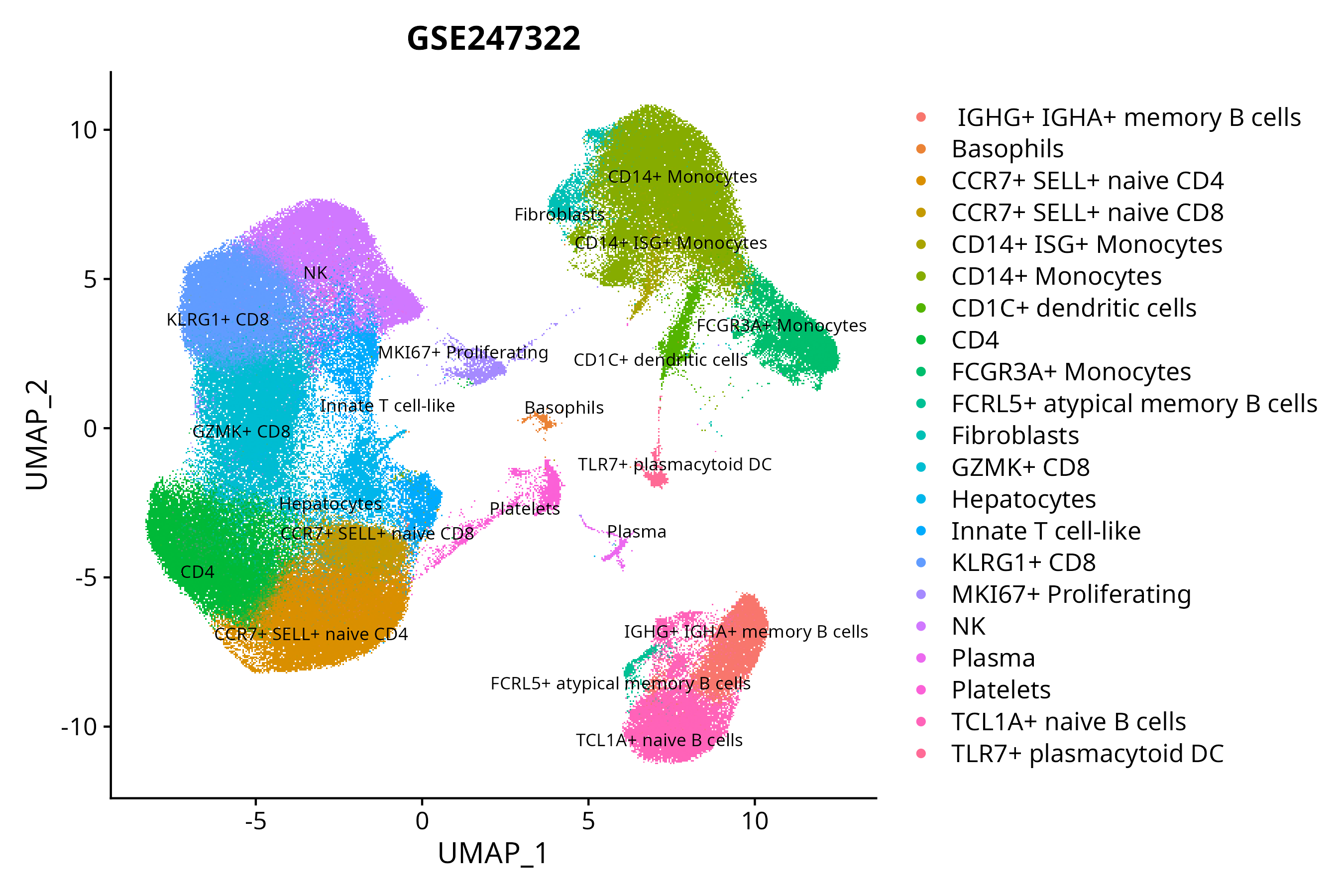
> Dataset: GSE247322 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information