Gene Information
|
Gene Name
|
CMAHP |
|
Gene ID
|
8418
|
|
Gene Full Name
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase, pseudogene |
|
Gene Alias
|
CMAH|CSAH |
|
Transcripts
|
ENSG00000293489
|
|
Virus
|
HPV |
|
Gene Type
|
pseudo |
|
HGNC ID
|
HGNC:2098
|
|
OMIM ID
|
603209 |
|
Summary
|
Sialic acids are terminal components of the carbohydrate chains of glycoconjugates involved in ligand-receptor, cell-cell, and cell-pathogen interactions. The two most common forms of sialic acid found in mammalian cells are N-acetylneuraminic acid (Neu5Ac) and its hydroxylated derivative, N-glycolylneuraminic acid (Neu5Gc). Studies of sialic acid distribution show that Neu5Gc is not detectable in normal human tissues although it was an abundant sialic acid in other mammals. Neu5Gc is, in actuality, immunogenic in humans. The absense of Neu5Gc in humans is due to a deletion within the human gene CMAH encoding cytidine monophosphate-N-acetylneuraminic acid hydroxylase, an enzyme responsible for Neu5Gc biosynthesis. Sequences encoding the mouse, pig, and chimpanzee hydroxylase enzymes were obtained by cDNA cloning and found to be highly homologous. However, the homologous human cDNA differs from these cDNAs by a 92-bp deletion in the 5' region. This deletion, corresponding to exon 6 of the mouse hydroxylase gene, causes a frameshift mutation and premature termination of the polypeptide chain in human. It seems unlikely that the truncated human hydroxylase mRNA encodes for an active enzyme explaining why Neu5Gc is undetectable in normal human tissues. Human genomic DNA also shows evidence of this deletion which does not occur in the genomes of African great apes. Nonetheless, the CMAH gene maps to 6p21.32 in humans and great apes indicating that mutation of the CMAH gene occurred following human divergence from chimpanzees and bonobos. [provided by RefSeq, Jul 2008] |
Target gene [CMAHP] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
Target gene [CMAHP] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE183048 - CMAHP peak across samples
|
Peak Plot
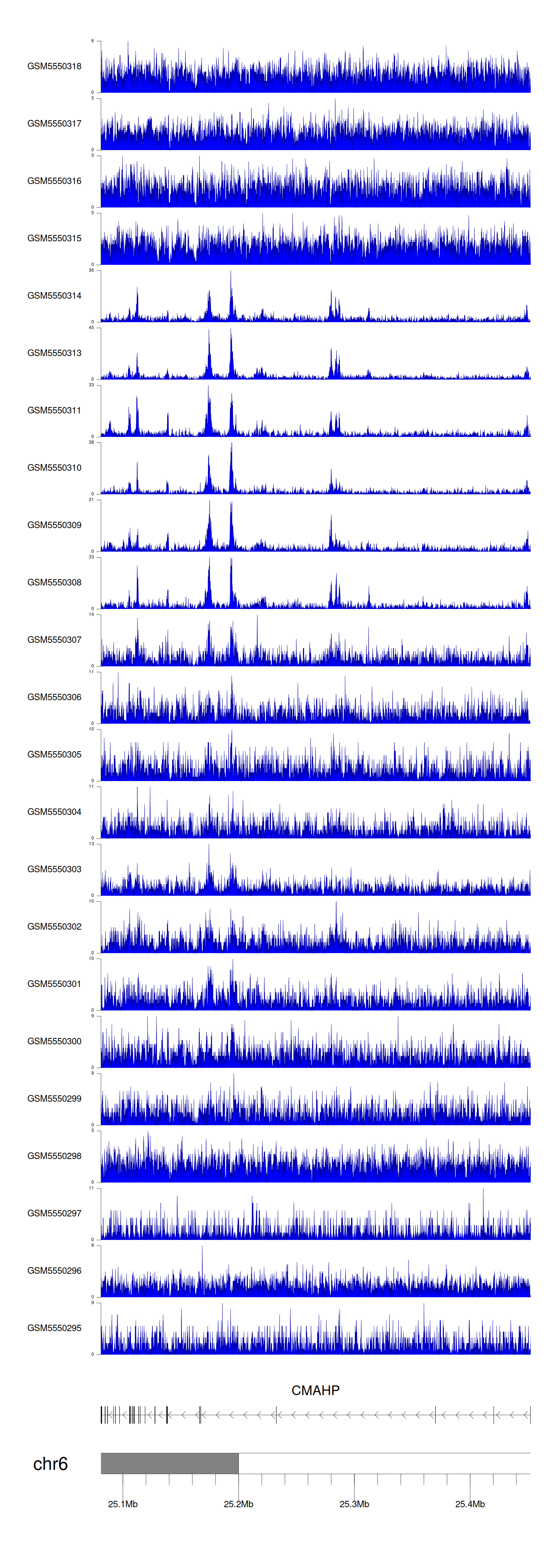
|
> Dataset: GSE143026 - CMAHP peak across samples
|
Peak Plot
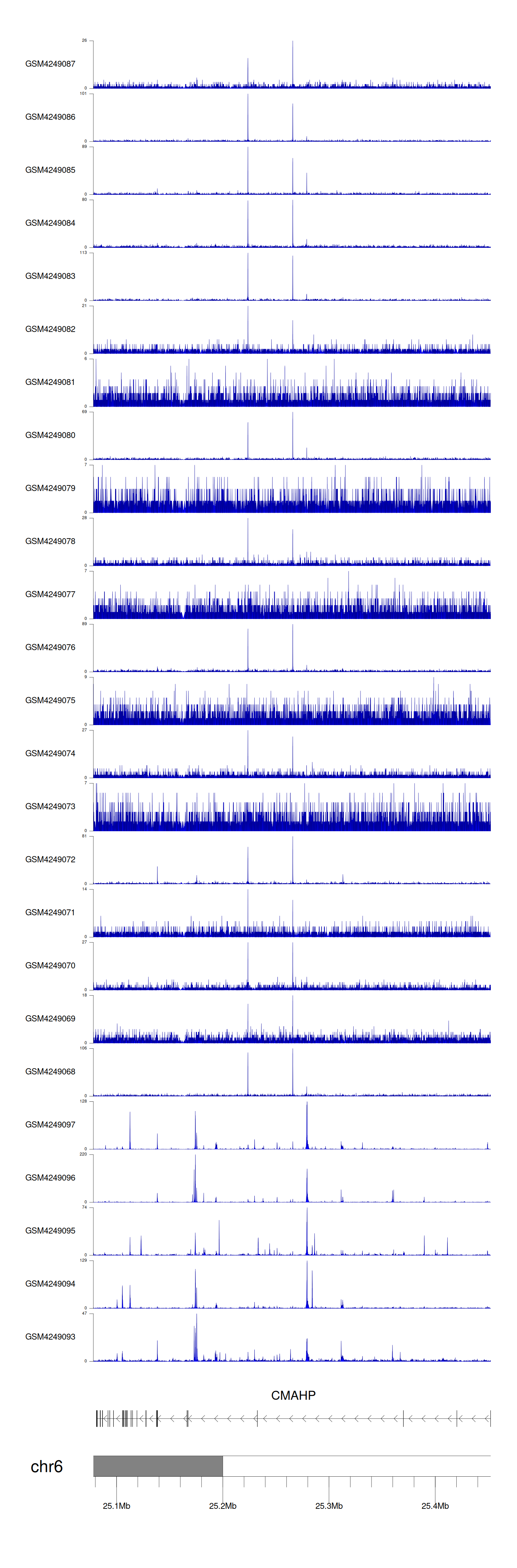
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information