Gene Information
|
Gene Name
|
FGF2 |
|
Gene ID
|
2247
|
|
Gene Full Name
|
fibroblast growth factor 2 |
|
Gene Alias
|
BFGF|FGF-2|FGFB|HBGF-2 |
|
Transcripts
|
ENSG00000138685
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm, Nuclear bodies;Secreted in other tissues; 
|
|
Membrane Info
|
Cancer-related genes, Candidate cardiovascular disease genes, FDA approved drug targets, Predicted intracellular proteins, Predicted secreted proteins, RAS pathway related proteins, Transporters |
|
Uniport_ID
|
P09038
|
|
HGNC ID
|
HGNC:3676
|
|
OMIM ID
|
134920 |
|
Summary
|
The protein encoded by this gene is a member of the fibroblast growth factor (FGF) family. FGF family members bind heparin and possess broad mitogenic and angiogenic activities. This protein has been implicated in diverse biological processes, such as limb and nervous system development, wound healing, and tumor growth. The mRNA for this gene contains multiple polyadenylation sites, and is alternatively translated from non-AUG (CUG) and AUG initiation codons, resulting in five different isoforms with distinct properties. The CUG-initiated isoforms are localized in the nucleus and are responsible for the intracrine effect, whereas, the AUG-initiated form is mostly cytosolic and is responsible for the paracrine and autocrine effects of this FGF. [provided by RefSeq, Jul 2008] |
Target gene [FGF2] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5010224 |
chr4 |
11 |
18 |
11 |
40 |
View |
| 5015958 |
chr4 |
13 |
0 |
0 |
13 |
View |
Target gene [FGF2] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE183048 - FGF2 peak across samples
|
Peak Plot
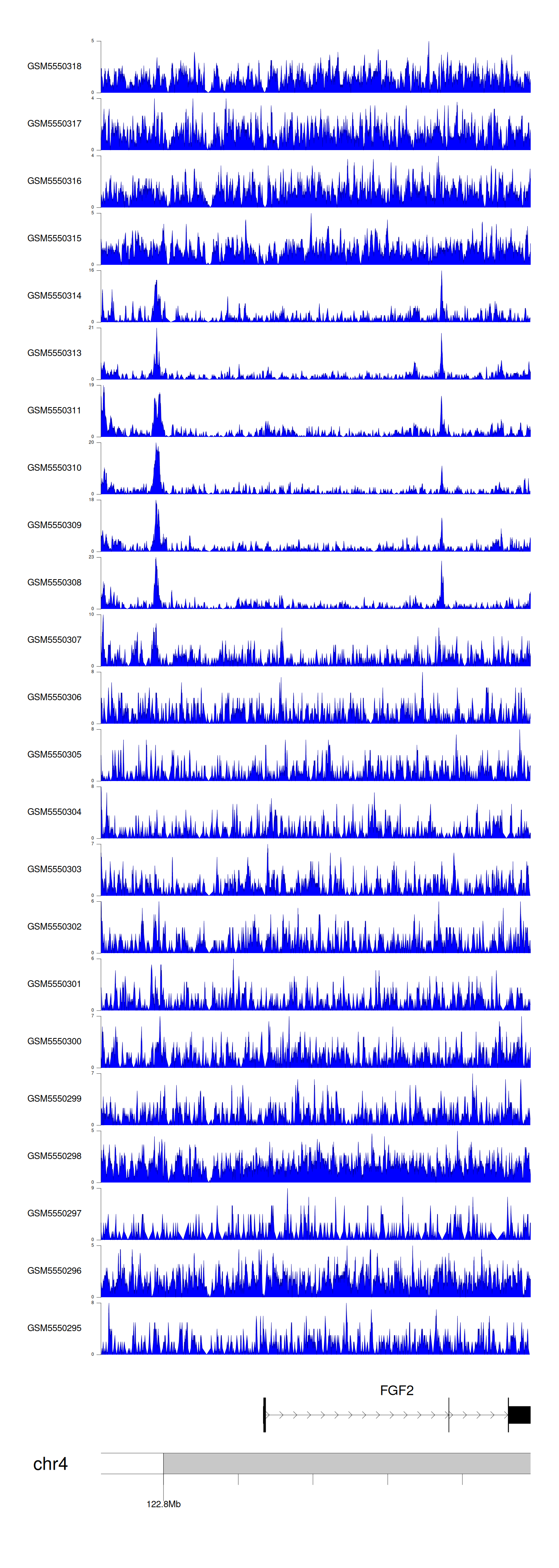
|
> Dataset: GSE143026 - FGF2 peak across samples
|
Peak Plot
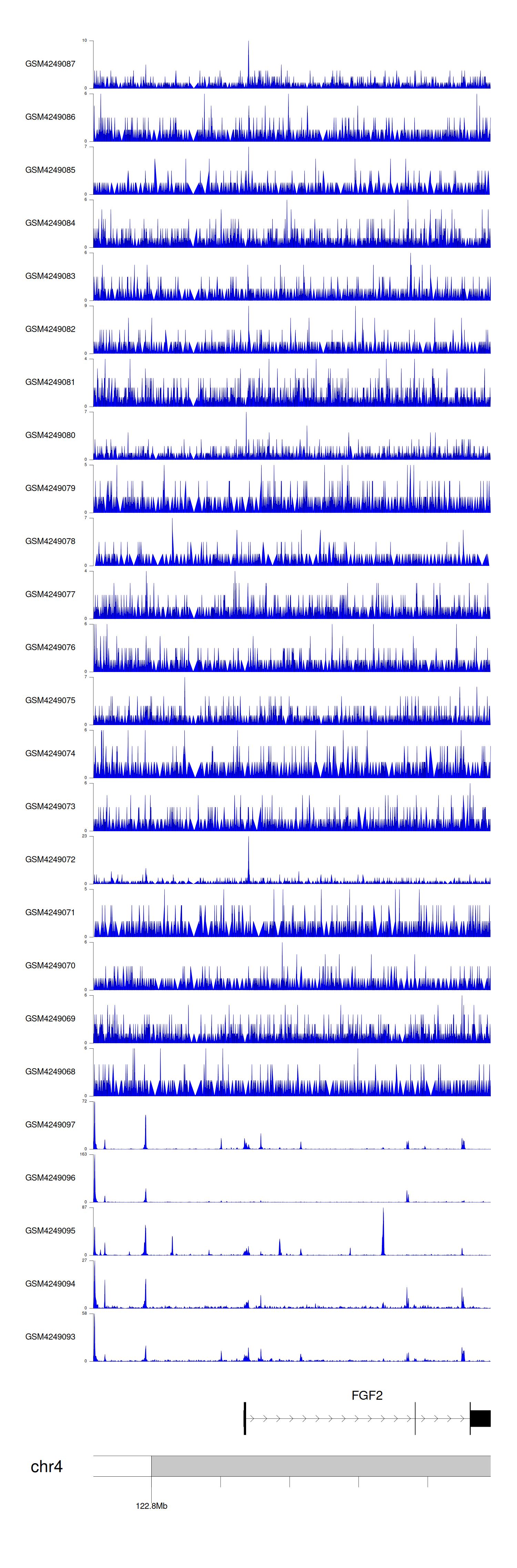
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information