Gene Information
|
Gene Name
|
KCNQ4 |
|
Gene ID
|
9132
|
|
Gene Full Name
|
potassium voltage-gated channel subfamily Q member 4 |
|
Gene Alias
|
DFNA2|DFNA2A|KV7.4 |
|
Transcripts
|
ENSG00000117013
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|

|
|
Membrane Info
|
Disease related genes, Human disease related genes, Potential drug targets, Predicted membrane proteins, Transporters, Voltage-gated ion channels |
|
Uniport_ID
|
P56696
|
|
HGNC ID
|
HGNC:6298
|
|
OMIM ID
|
603537 |
|
Summary
|
The protein encoded by this gene forms a potassium channel that is thought to play a critical role in the regulation of neuronal excitability, particularly in sensory cells of the cochlea. The current generated by this channel is inhibited by M1 muscarinic acetylcholine receptors and activated by retigabine, a novel anti-convulsant drug. The encoded protein can form a homomultimeric potassium channel or possibly a heteromultimeric channel in association with the protein encoded by the KCNQ3 gene. Defects in this gene are a cause of nonsyndromic sensorineural deafness type 2 (DFNA2), an autosomal dominant form of progressive hearing loss. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008] |
Target gene [KCNQ4] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5012324 |
chr1 |
9 |
3 |
0 |
12 |
View |
| 5012325 |
chr1 |
39 |
1 |
0 |
40 |
View |
Target gene [KCNQ4] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE143026 - KCNQ4 peak across samples
|
Peak Plot
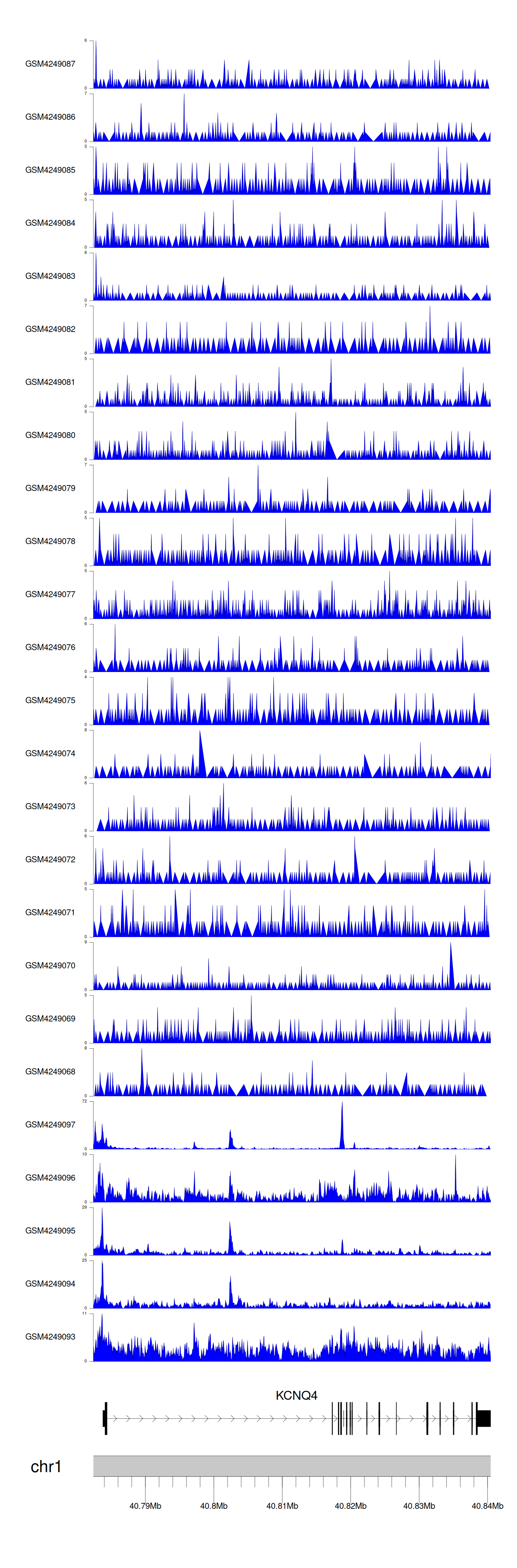
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information