Gene Information
|
Gene Name
|
RAP1GAP |
|
Gene ID
|
5909
|
|
Gene Full Name
|
RAP1 GTPase activating protein |
|
Gene Alias
|
RAP1GA1|RAP1GAP1|RAP1GAPII|RAPGAP |
|
Transcripts
|
ENSG00000076864
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Cytosol; 
|
|
Membrane Info
|
Predicted intracellular proteins |
|
Uniport_ID
|
P47736
|
|
HGNC ID
|
HGNC:9858
|
|
OMIM ID
|
600278 |
|
Summary
|
This gene encodes a type of GTPase-activating-protein (GAP) that down-regulates the activity of the ras-related RAP1 protein. RAP1 acts as a molecular switch by cycling between an inactive GDP-bound form and an active GTP-bound form. The product of this gene, RAP1GAP, promotes the hydrolysis of bound GTP and hence returns RAP1 to the inactive state whereas other proteins, guanine nucleotide exchange factors (GEFs), act as RAP1 activators by facilitating the conversion of RAP1 from the GDP- to the GTP-bound form. In general, ras subfamily proteins, such as RAP1, play key roles in receptor-linked signaling pathways that control cell growth and differentiation. RAP1 plays a role in diverse processes such as cell proliferation, adhesion, differentiation, and embryogenesis. Alternative splicing results in multiple transcript variants encoding distinct proteins. [provided by RefSeq, Aug 2011] |
Target gene [RAP1GAP] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5016928 |
chr1 |
10 |
10 |
2 |
22 |
View |
Target gene [RAP1GAP] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
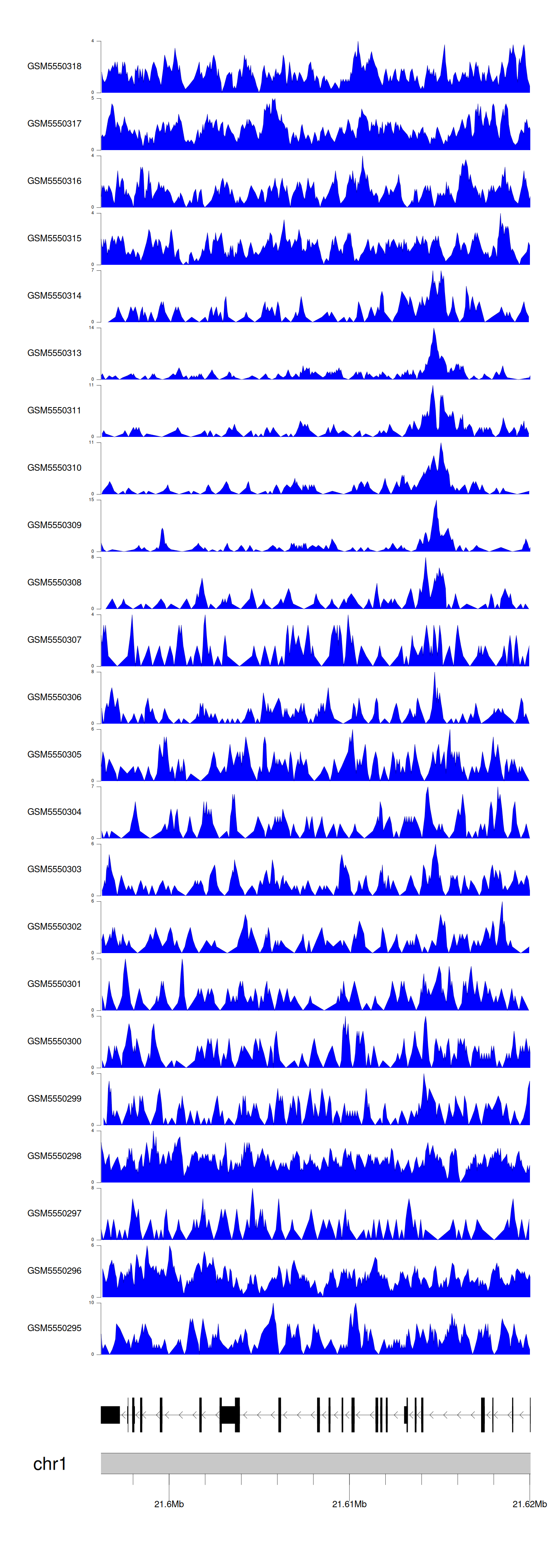
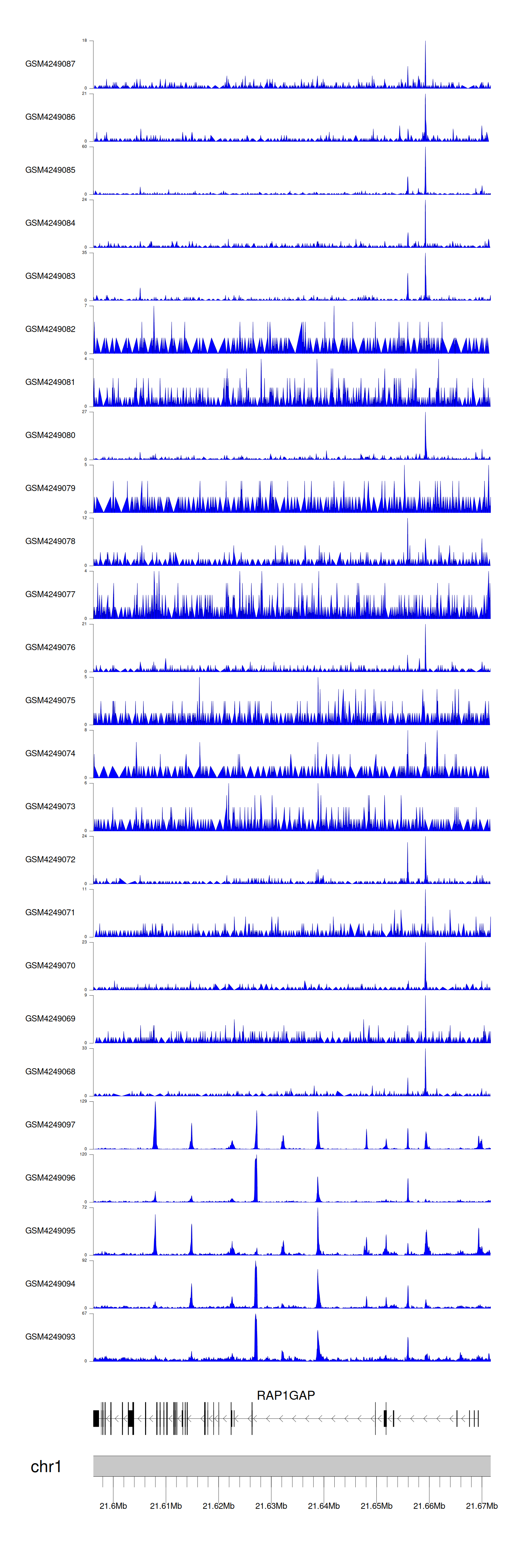
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE183048 - RAP1GAP peak across samples
|
Peak Plot
|
> Dataset: GSE143026 - RAP1GAP peak across samples
|
Peak Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information