Gene Information
|
Gene Name
|
CTSL |
|
Gene ID
|
1514
|
|
Gene Full Name
|
cathepsin L |
|
Gene Alias
|
CATL|CTSL1|MEP |
|
Transcripts
|
ENSG00000135047
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Golgi apparatus, Vesicles;Intracellular and membrane; 
|
|
Membrane Info
|
Cancer-related genes, Enzymes, Metabolic proteins, Plasma proteins, Predicted intracellular proteins |
|
Uniport_ID
|
P07711
|
|
HGNC ID
|
HGNC:2537
|
|
OMIM ID
|
116880 |
|
Summary
|
The protein encoded by this gene is a lysosomal cysteine proteinase that plays a major role in intracellular protein catabolism. Its substrates include collagen and elastin, as well as alpha-1 protease inhibitor, a major controlling element of neutrophil elastase activity. The encoded protein has been implicated in several pathologic processes, including myofibril necrosis in myopathies and in myocardial ischemia, and in the renal tubular response to proteinuria. This protein, which is a member of the peptidase C1 family, is a dimer composed of disulfide-linked heavy and light chains, both produced from a single protein precursor. Additionally, this protein cleaves the S1 subunit of the SARS-CoV-2 spike protein, which is necessary for entry of the virus into the cell. [provided by RefSeq, Aug 2020] |
Target gene [CTSL] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5016465 |
chr9 |
97 |
7 |
0 |
104 |
View |
Target gene [CTSL] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
S GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
S GSE139324
|
scRNA-seq |
63 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
S GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
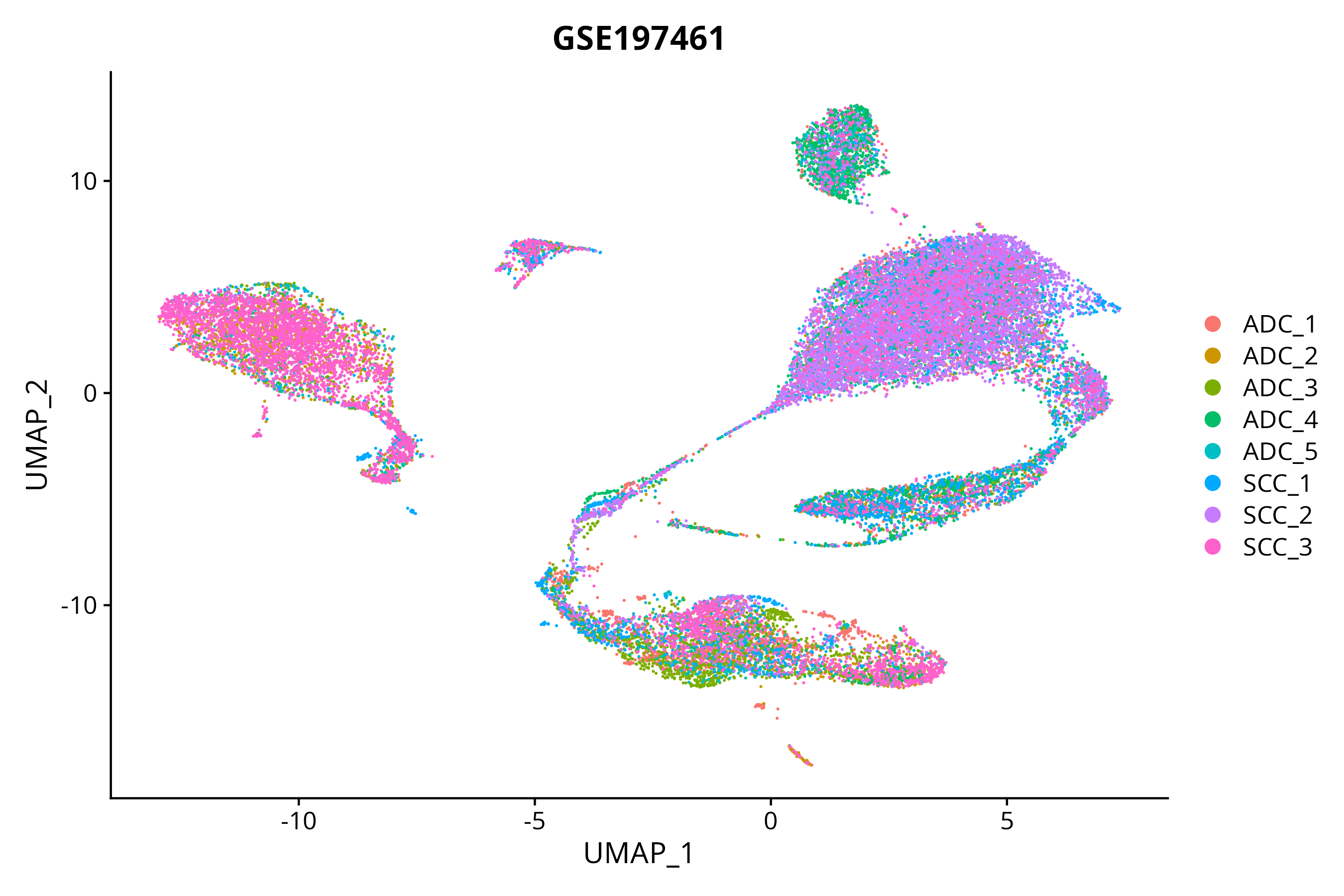
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
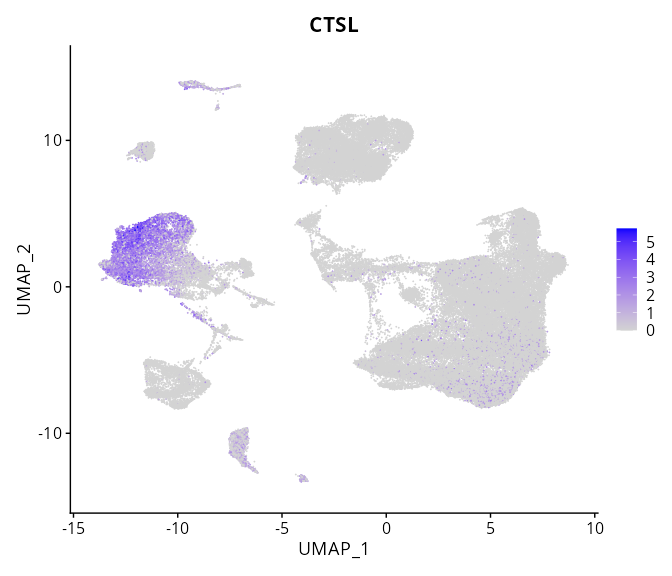
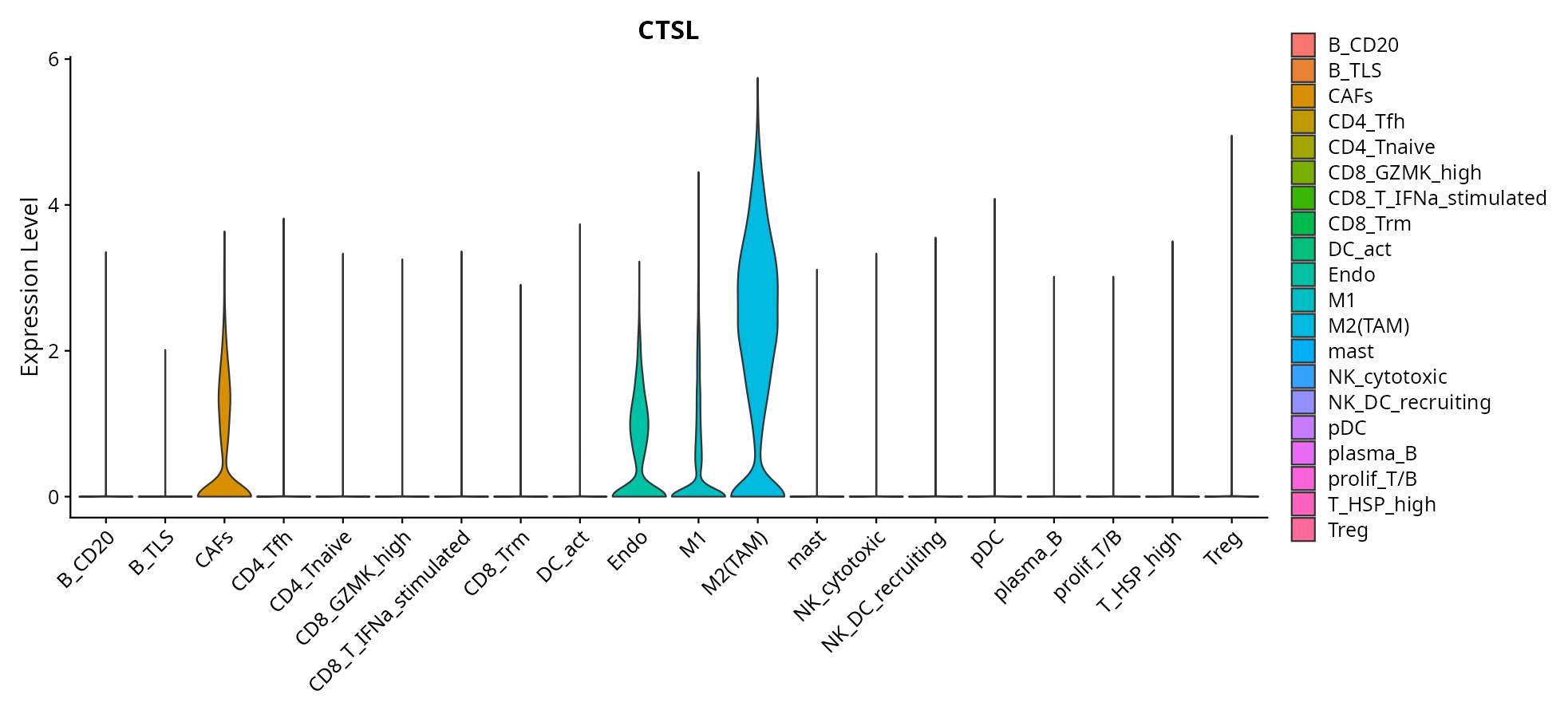
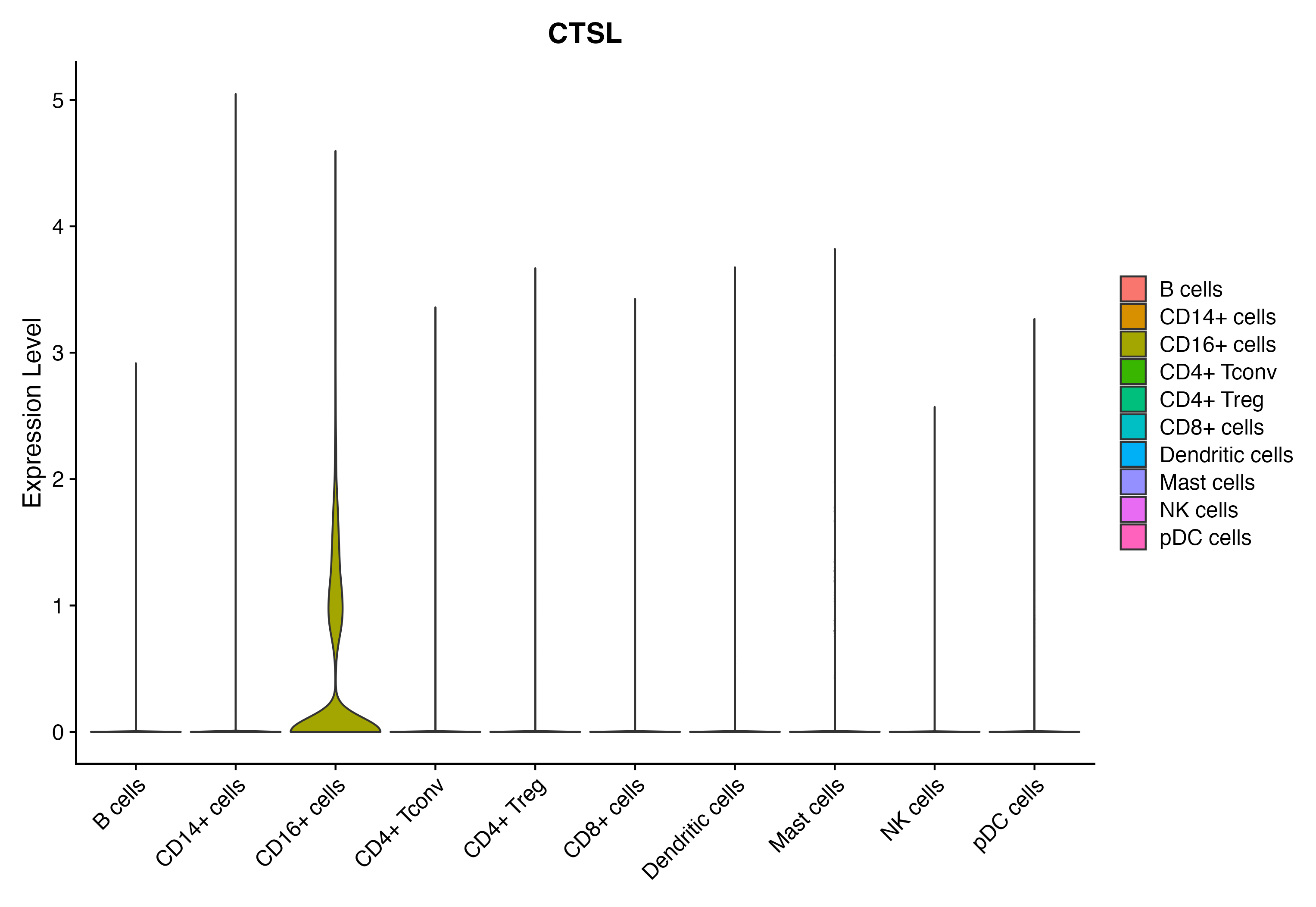
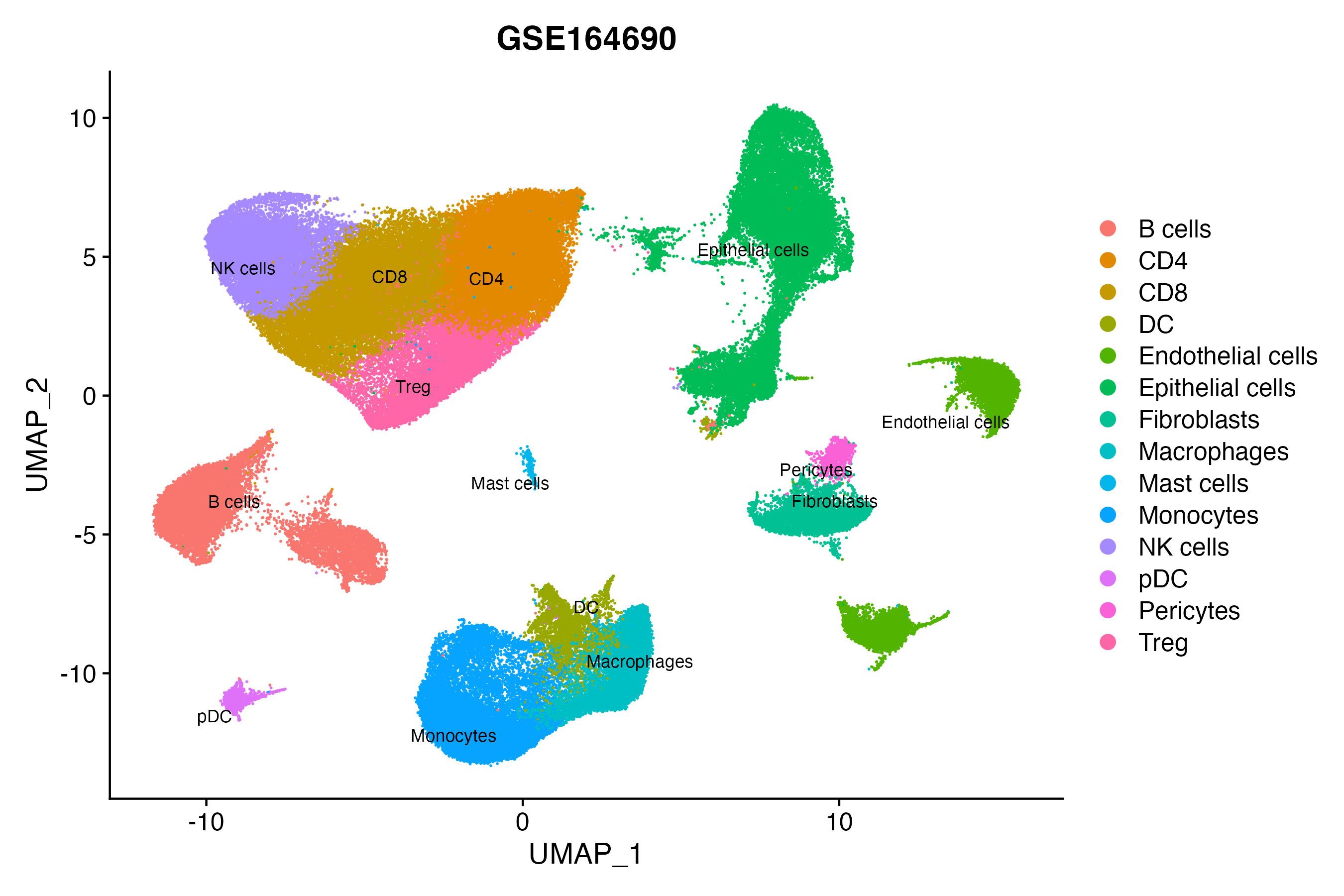
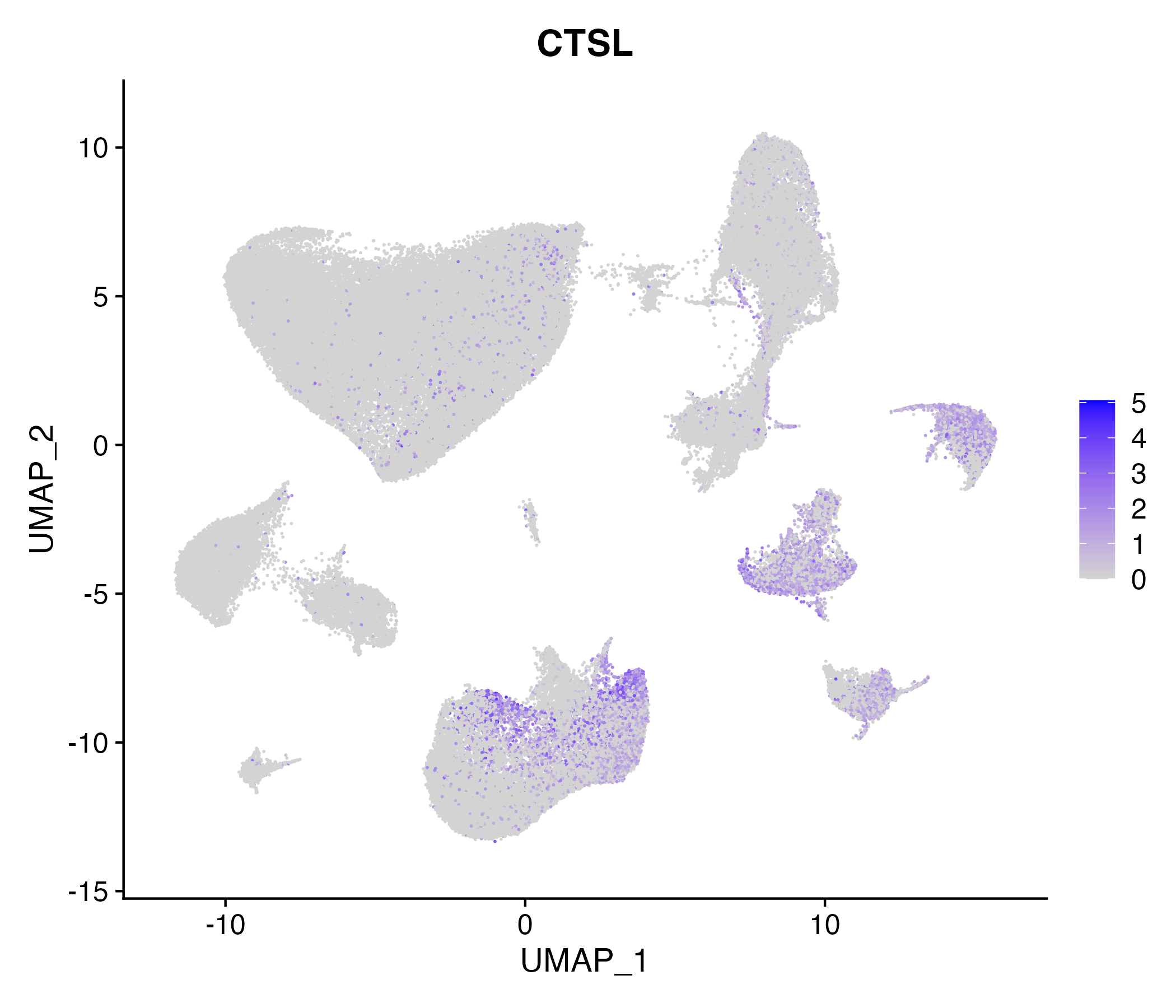
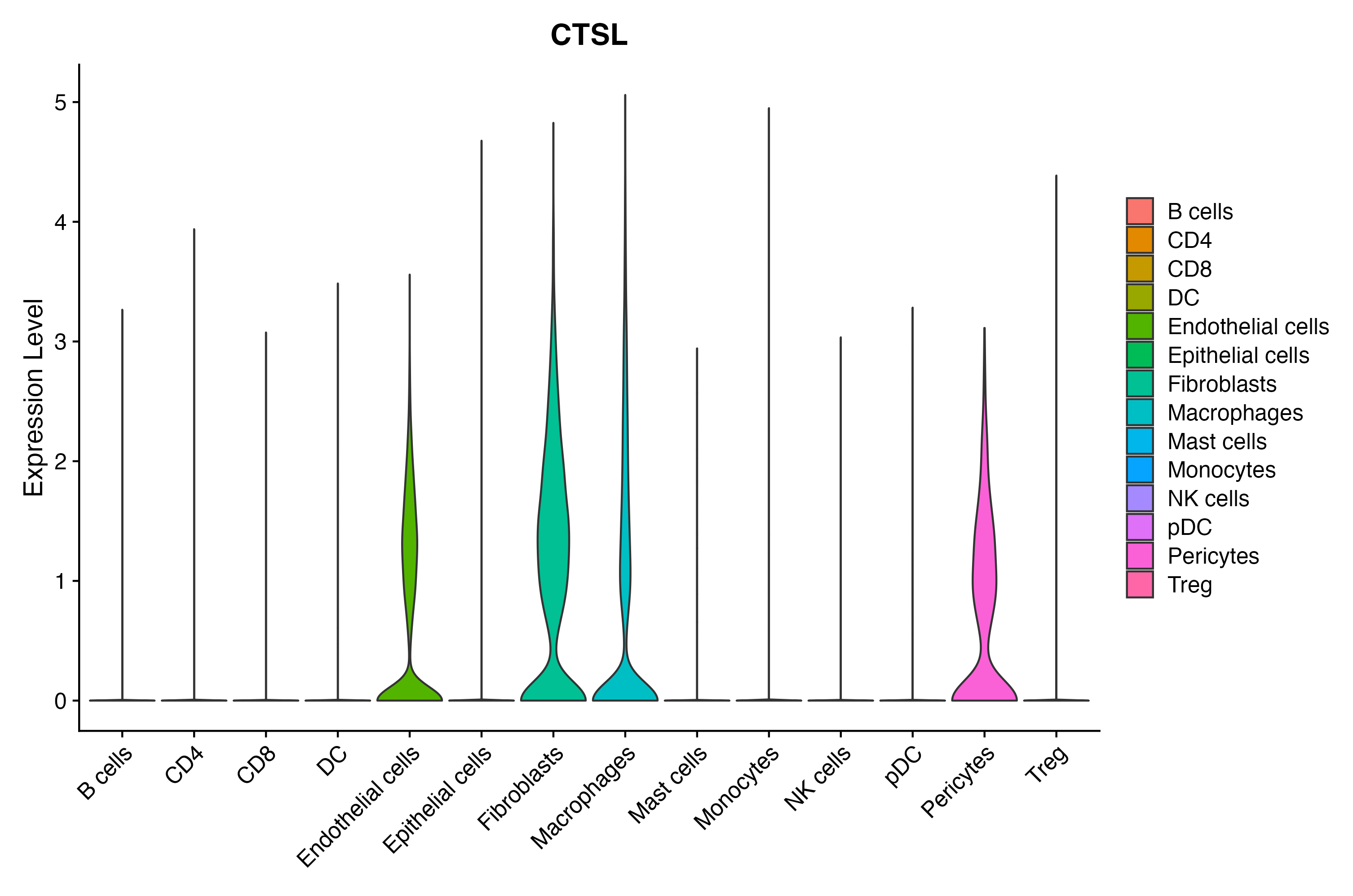
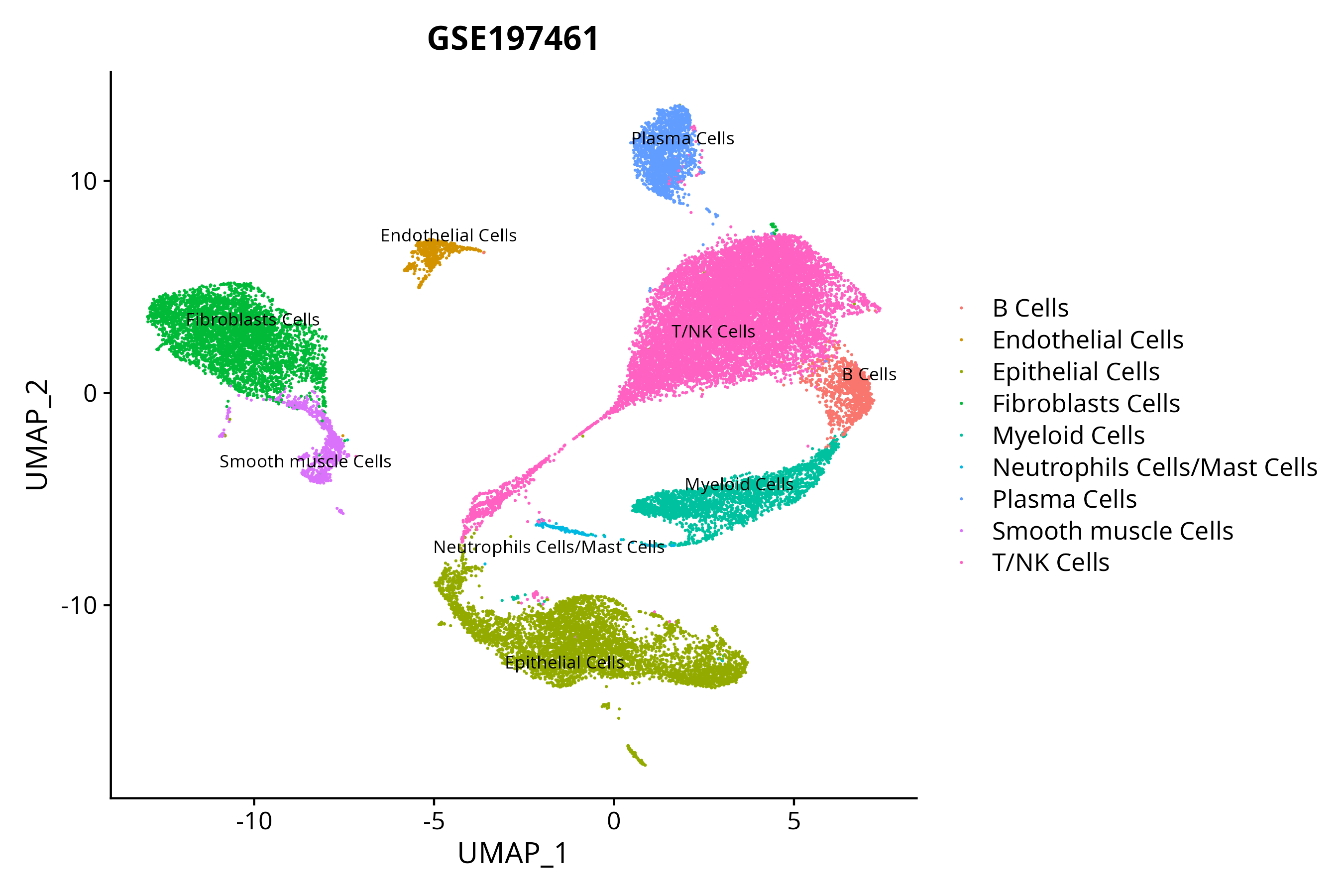
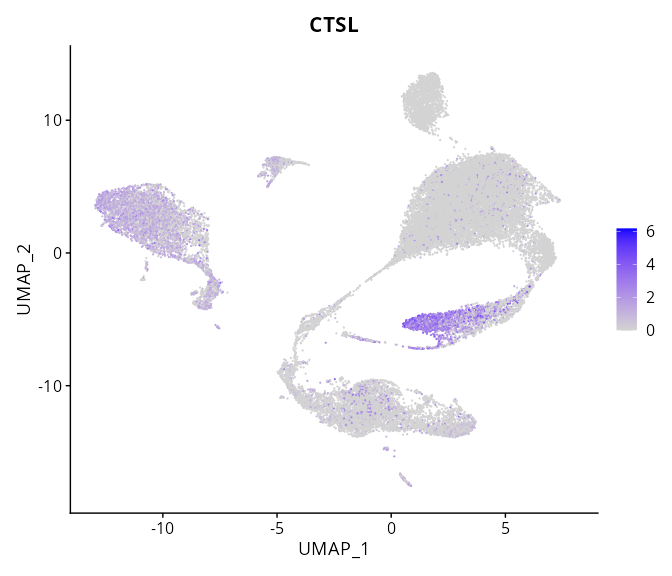
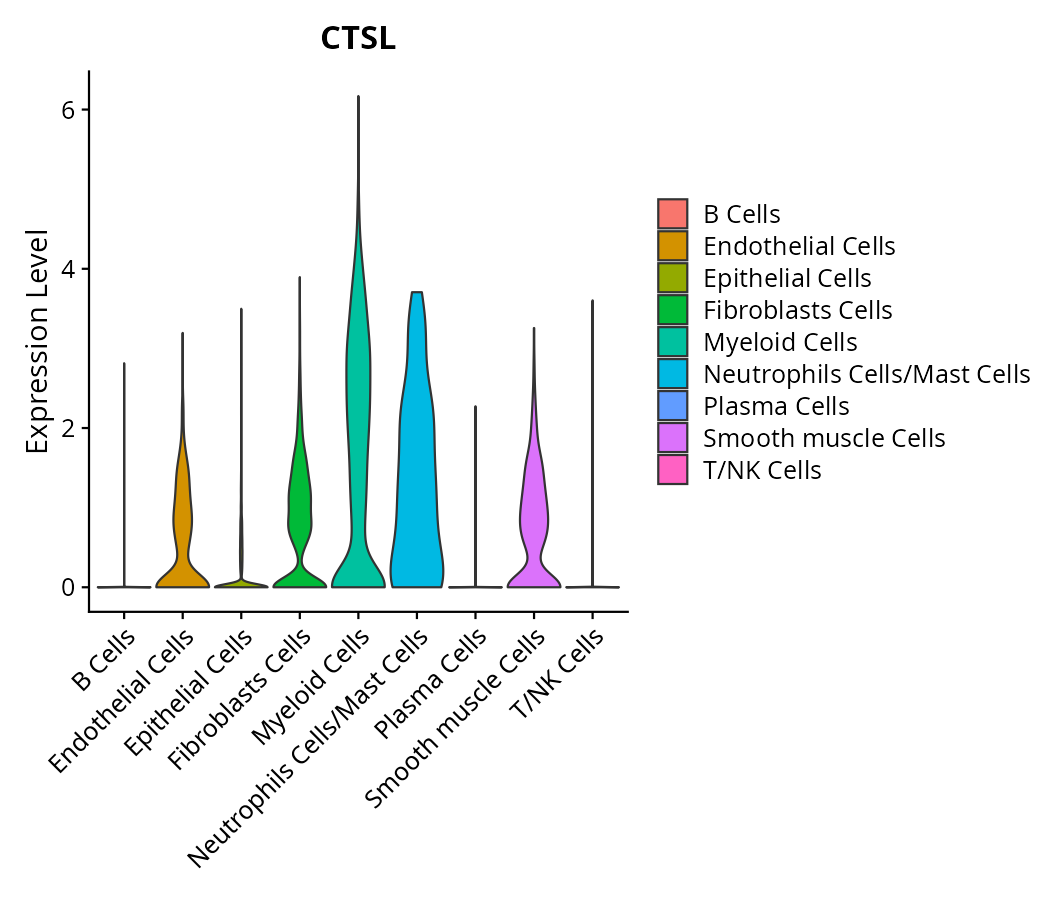
When the query gene is differentially changed in the dataset, a feature/violin plot will be displayed.
> Dataset: GSE226620 - Gene expression in cell subsets
|
|
|

|
|
|
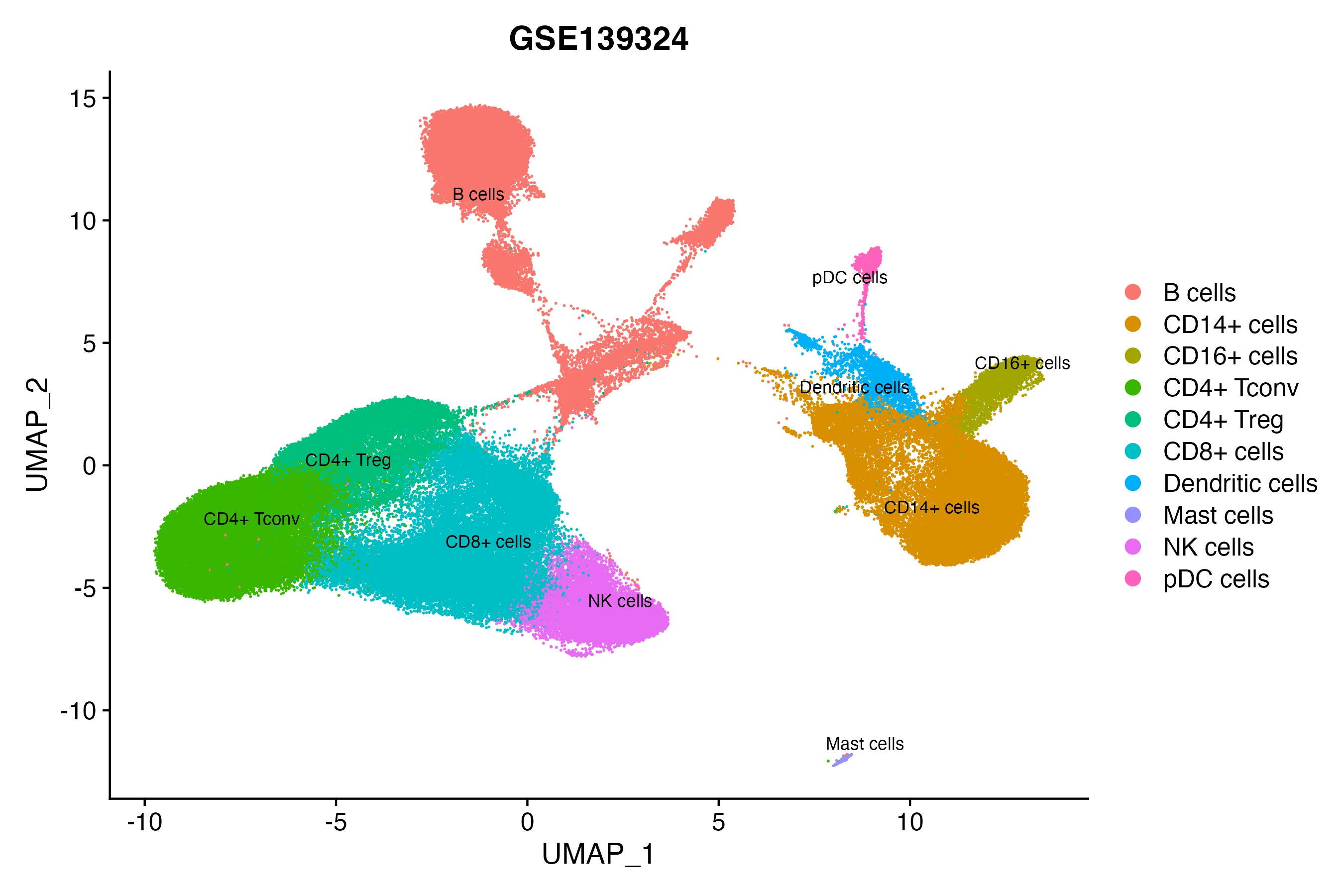
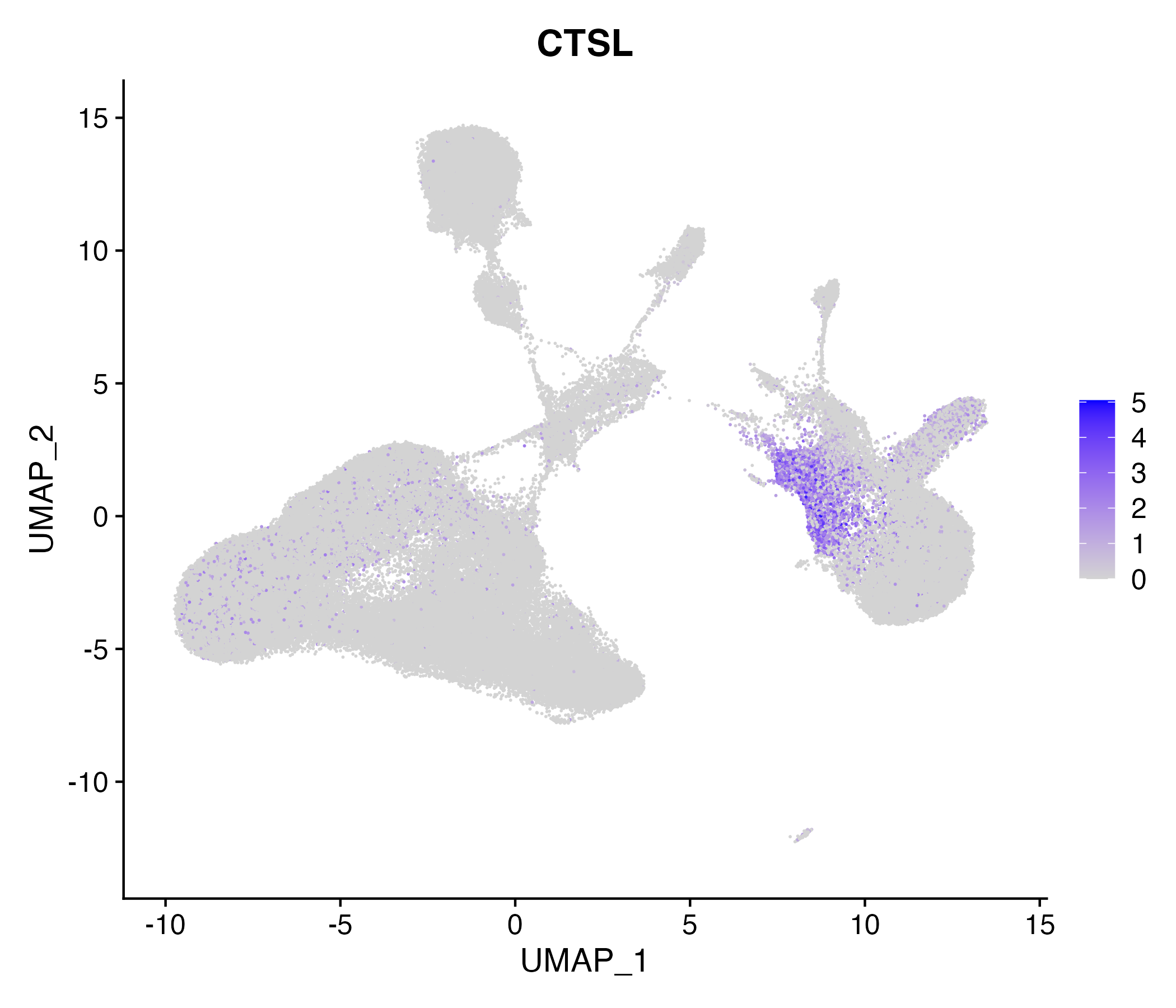
> Dataset: GSE139324 - Gene expression in cell subsets
|
|
|
|
|
|
> Dataset: GSE164690 - Gene expression in cell subsets
|
|
|

|
|
|
> Dataset: GSE197461 - Gene expression in cell subsets
|
|
|
|
|
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information