Gene Information
|
Gene Name
|
ERG |
|
Gene ID
|
2078
|
|
Gene Full Name
|
ETS transcription factor ERG |
|
Gene Alias
|
LMPHM14|erg-3|p55 |
|
Transcripts
|
ENSG00000157554
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Cytosol; 
|
|
Membrane Info
|
Cancer-related genes, Disease related genes, Human disease related genes, Predicted intracellular proteins, Transcription factors |
|
Uniport_ID
|
P11308
|
|
HGNC ID
|
HGNC:3446
|
|
OMIM ID
|
165080 |
|
Summary
|
This gene encodes a member of the erythroblast transformation-specific (ETS) family of transcriptions factors. All members of this family are key regulators of embryonic development, cell proliferation, differentiation, angiogenesis, inflammation, and apoptosis. The protein encoded by this gene is mainly expressed in the nucleus. It contains an ETS DNA-binding domain and a PNT (pointed) domain which is implicated in the self-association of chimeric oncoproteins. This protein is required for platelet adhesion to the subendothelium, inducing vascular cell remodeling. It also regulates hematopoesis, and the differentiation and maturation of megakaryocytic cells. This gene is involved in chromosomal translocations, resulting in different fusion gene products, such as TMPSSR2-ERG and NDRG1-ERG in prostate cancer, EWS-ERG in Ewing's sarcoma and FUS-ERG in acute myeloid leukemia. More than two dozens of transcript variants generated from combinatorial usage of three alternative promoters and multiple alternative splicing events have been reported, but the full-length nature of many of these variants has not been determined. [provided by RefSeq, Apr 2014] |
Target gene [ERG] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5005367 |
chr21 |
23 |
34 |
0 |
57 |
View |
| 5007312 |
chr21 |
9 |
0 |
0 |
9 |
View |
| 5010321 |
chr21 |
1 |
0 |
0 |
1 |
View |
| 5011466 |
chr21 |
26 |
11 |
0 |
37 |
View |
Target gene [ERG] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
S GSE226620
|
scRNA-seq |
30 |
HiSeq X Ten (Homo sapiens) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
S GSE164690
|
scRNA-seq |
51 |
Illumina NextSeq 500 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
E GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
M TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
S GSE197461
|
scRNA-seq |
16 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the query gene is differentially changed in the dataset, a volcano/bar plot will be displayed.
> Dataset: GSE55542 - ERG expression across samples
|
Volcano Plot
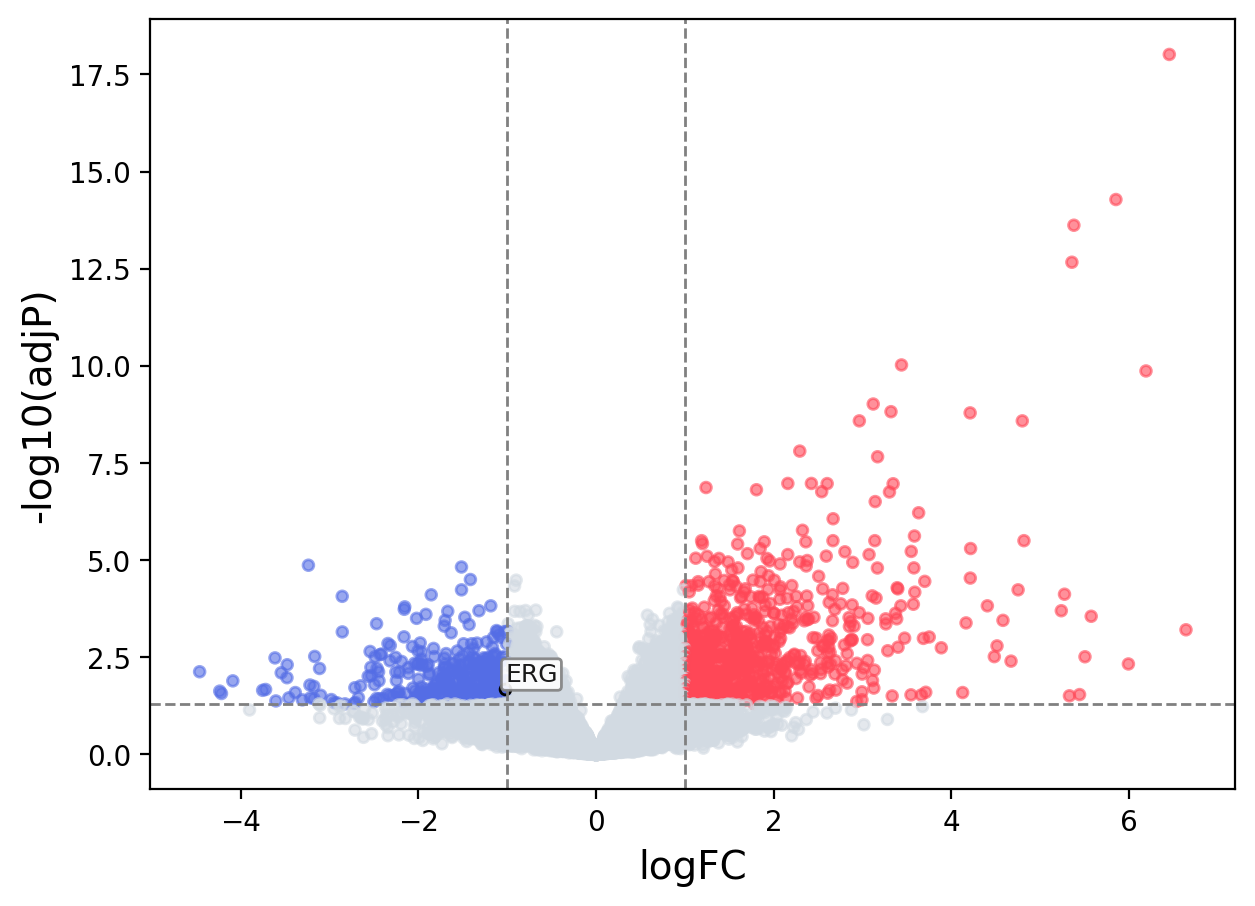
|
Bar Plot
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information