Gene Information
|
Gene Name
|
OAZ3 |
|
Gene ID
|
51686
|
|
Gene Full Name
|
ornithine decarboxylase antizyme 3 |
|
Gene Alias
|
AZ3|OAZ-t|TISP15 |
|
Transcripts
|
ENSG00000143450
|
|
Virus
|
HPV |
|
Gene Type
|
protein-coding |
|
HPA Location Info
|
Nucleoplasm;Mid piece, Principal piece; 
|
|
Membrane Info
|
Predicted intracellular proteins |
|
Uniport_ID
|
Q9UMX2
|
|
HGNC ID
|
HGNC:8097
|
|
OMIM ID
|
605138 |
|
Summary
|
The protein encoded by this gene belongs to the ornithine decarboxylase antizyme family, which plays a role in cell growth and proliferation by regulating intracellular polyamine levels. Expression of antizymes requires +1 ribosomal frameshifting, which is enhanced by high levels of polyamines. Antizymes in turn bind to and inhibit ornithine decarboxylase (ODC), the key enzyme in polyamine biosynthesis; thus, completing the auto-regulatory circuit. This gene encodes antizyme 3, the third member of the antizyme family. Like antizymes 1 and 2, antizyme 3 inhibits ODC activity and polyamine uptake; however, it does not stimulate ODC degradation. Also, while antizymes 1 and 2 have broad tissue distribution, expression of antizyme 3 is restricted to haploid germ cells in testis, suggesting a distinct role for this antizyme in spermiogenesis. Antizyme 3 gene knockout studies showed that homozygous mutant male mice were infertile, and indicated the likely role of this antizyme in the formation of a rigid connection between the sperm head and tail during spermatogenesis. Alternatively spliced transcript variants encoding different isoforms, including one resulting from the use of non-AUG (CUG) translation initiation codon, have been found for this gene. [provided by RefSeq, Dec 2014] |
Target gene [OAZ3] related to VISs 
Integration Table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in this section
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 5014629 |
chr1 |
4400 |
2543 |
1172 |
8115 |
View |
Target gene [OAZ3] related to Omics data
| Data ID |
Experiment type |
Sample number |
Platform |
|
C GSE183048
|
Chip-seq |
24 |
Illumina HiSeq 4000 (Homo sapiens) |
|
GSE40774
|
Expression array |
134 |
Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version) |
|
GSE169622
|
Methylation profiling (Array) |
9 |
Infinium MethylationEPIC |
|
GSE65858
|
Expression array |
270 |
Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE181805
|
Expression array |
25 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE196215
|
RNA-seq |
8 |
Illumina NovaSeq 6000 (Homo sapiens) |
|
GSE138080
|
Expression array |
35 |
Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version) |
|
GSE140662
|
Expression array |
8 |
[HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
|
GSE55542
|
Expression array |
36 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
C GSE143026
|
ATAC-seq;Chip-seq;RNA-seq |
30 |
Illumina HiSeq 2500 (Homo sapiens) |
|
TCGA_CESC
|
DNA methylation sequencing;RNA-seq |
288 |
TCGA |
|
GSE51993
|
Expression array |
48 |
Illumina Human v2 MicroRNA expression beadchip;Illumina HumanHT-12 V4.0 expression beadchip |
|
GSE55550
|
Expression array |
155 |
Agilent-039494 SurePrint G3 Human GE v2 8x60K Microarray 039381 (Probe Name version) |
|
GSE165883
|
RNA-seq |
20 |
Illumina NextSeq 500 (Homo sapiens) |
When the gene can detect a peak in the dataset, a peak plot will be displayed.
> Dataset: GSE183048 - OAZ3 peak across samples
|
Peak Plot
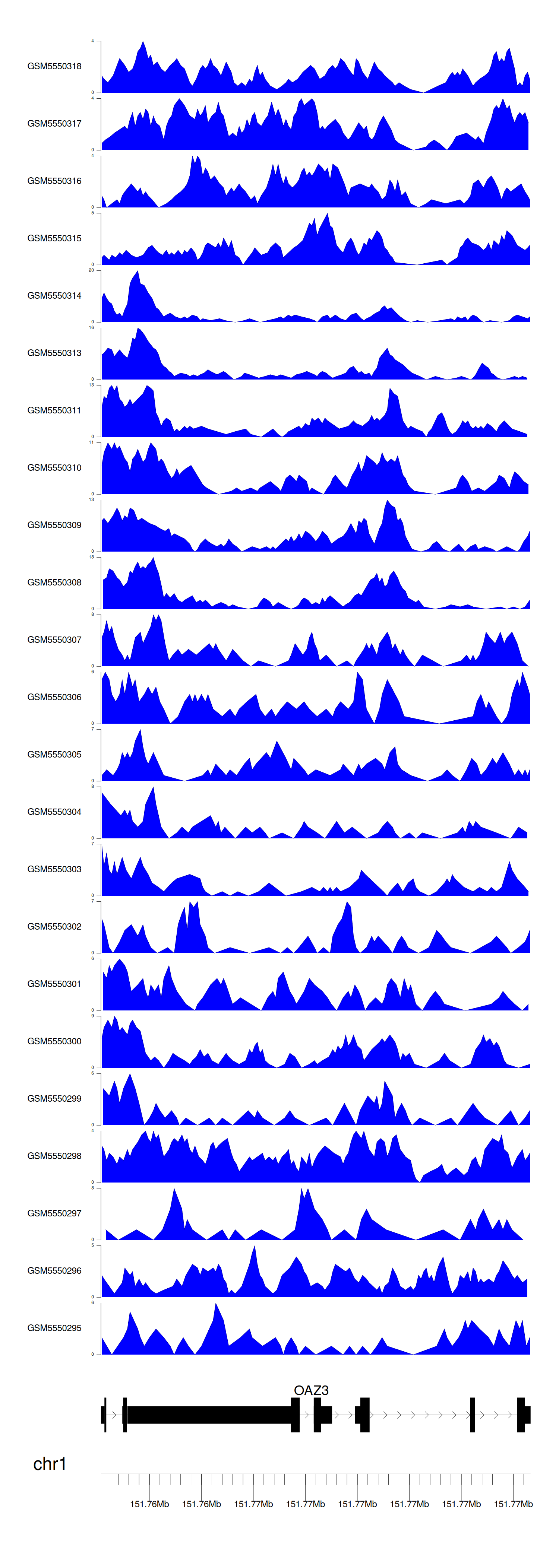
|
> Dataset: GSE143026 - OAZ3 peak across samples
|
Peak Plot
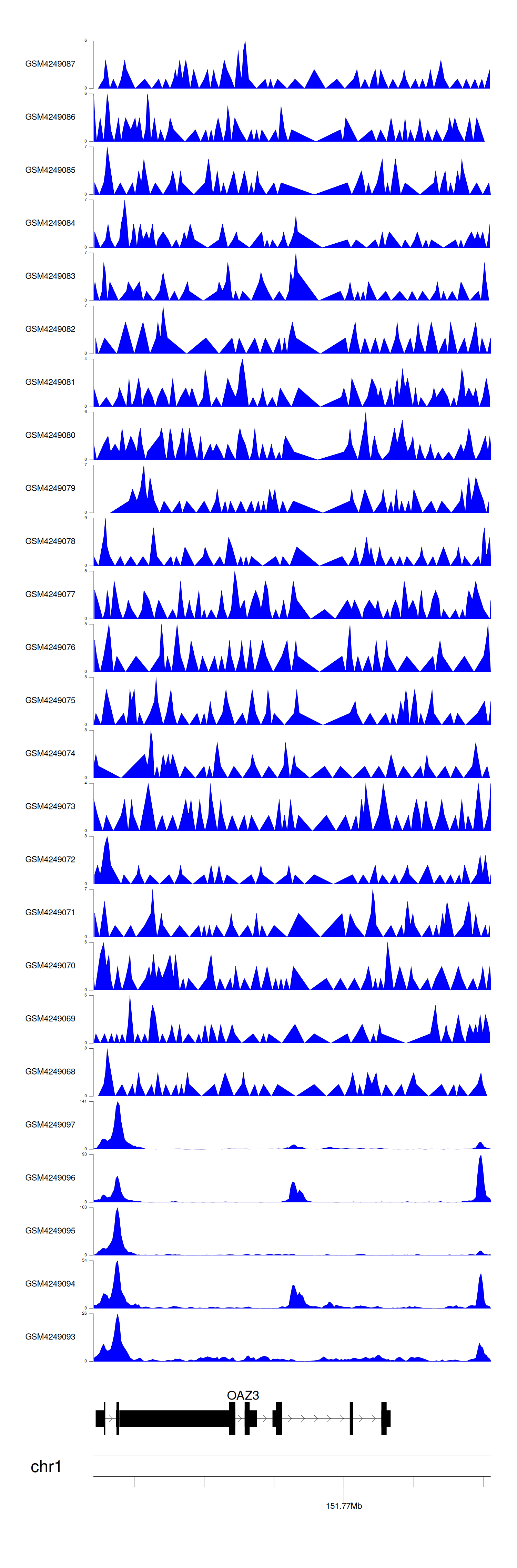
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information